
Research strategy and structure of the CRC
(1) Interdisciplinary tandem projects for tool development breakthroughs.
(2) Project areas.
(3) Cross-area interactions and replication-cycle coverage.
Many of the CRC’s overarching research questions listed on require integration across all three project areas. These questions collectively span all steps of the virus replication cycle, Fig. 7, from genome replication and transcription to virus assembly, entry, tropism, and host response. In addition, the CRC addresses complementary aspects such as phylogeny, quasispecies dynamics, intravirus interactions, virus particle sorting, surveillance, and vaccine design, ensuring a comprehensive approach to virus characterisation and risk assessment. By combining tools from the three project areas, the consortium will integrate sequence, molecular, structural, and phenotypic data to create comprehensive models of virus biology. This will enable the CRC to generate new insights, develop predictive approaches, and directly support the overarching aim of virus risk assessment.
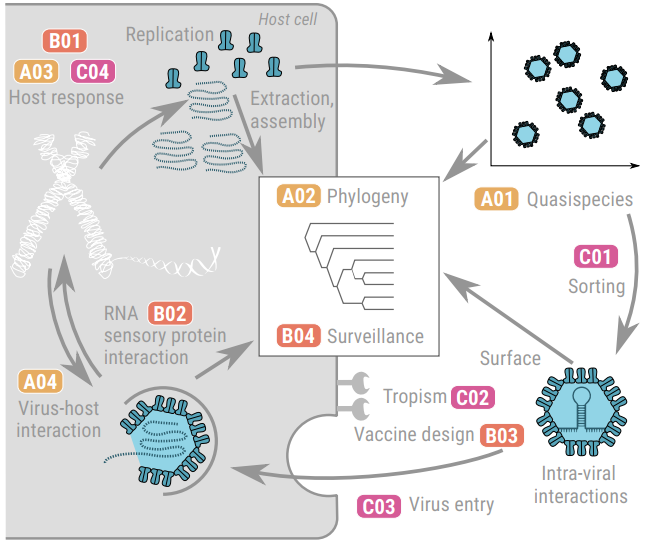
Overview of the individual projects and their interaction within the CRC VirusREvolution.
