
Overarching research goal of the CRC VirusREvolution
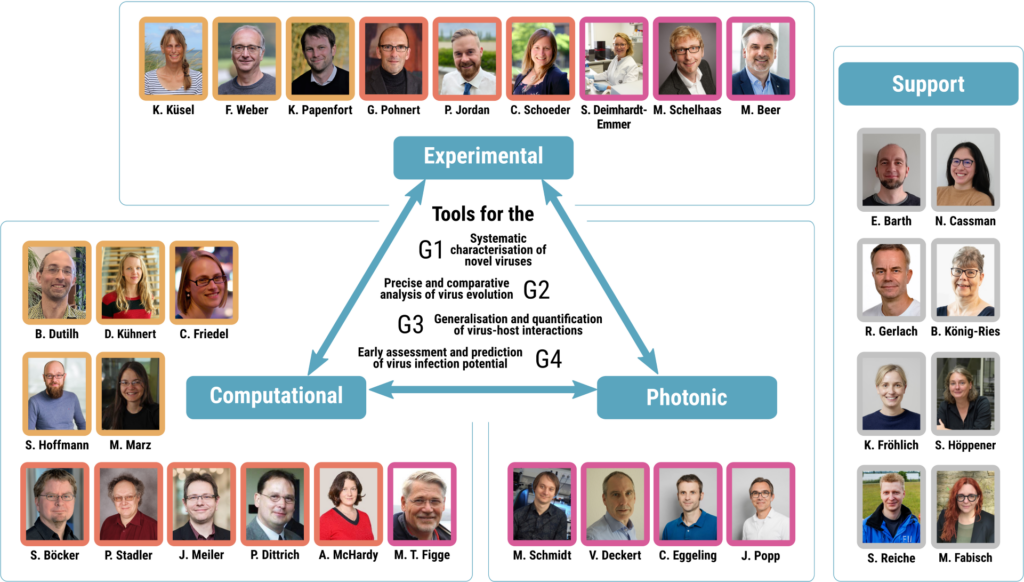
The primary objective of VirusREvolution is to pioneer the creation of specialized bioinformatic and photonic tools dedicated to viruses. More specifically we have four different goals:
G1 Tools for the systematic characterisation of novel viruses.
It is of utmost importance to detect and characterise fast and efficiently novel emerging viruses. Within VirusREvolution we will develop tools for virus identification, annotation, alignments and general characterisation on the level of genome and morphology.
G2 Tools for precise and comparative analysis of virus evolution.
We will develop tools for phylogeny of viruses on two different levels: we will improve the overall virus long-term phylogeny by precise annotation of viruses (macroevolution) and we will describe the quasispecies and short-term surveillance of a virus (microevolution).
G3 Tools for the generalisation and quantification of virus-host interactions.
We will analyse the transcriptome, metabolome, proteome and their correlation. Therefore, we will strengthen the first insights into the virus-induced host degradosome and the functionality of enzymes. Additionally, we aim to understand natural intracellular and epicellular anti-viral strategies and leverage the knowledge to design improved antiviral reagents.
G4 Tools for the early assessment and prediction of virus infection potential.
Using a broad range of optimised and tailored complementary tools, we aim to disclose and further understand virus characteristics, such as virus-cell entry and uncoating mechanisms, to predict pathogenicity and tropism and to use selected features for virus sorting.
