

High-resolution and high-throughput optical detection and characterisation of viruses
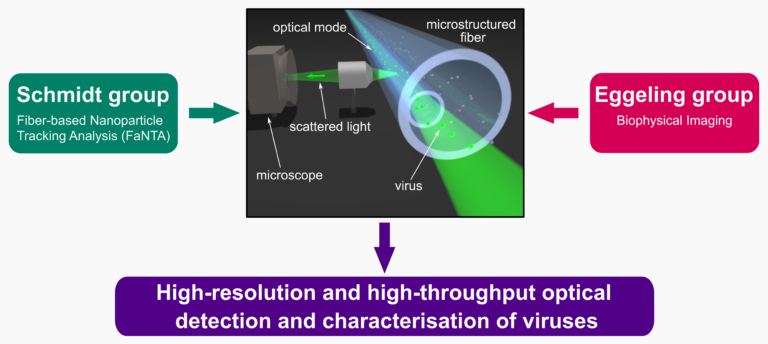
Here, the structural parameters of virus particles diffusing through a detection spot in an optical fiber or waveguide will be retrieved using measurements of elastic scattering and/or fluorescence emission. In this, the most challenging issues are the distinguishing of different viruses and of unwanted background material, which will be approached through optimisation of the fibers, correlation of readouts, and tailored data analysis. Specifically, we aim to employ a combination of label-free scattering-based detection and generic fluorescence labelling approaches, novel fiber technology, advanced scattering approaches, and fluorescence spectroscopy, and their joint detection and analysis. Analysis specifically includes a tailored interpretation of diffusional tracks, and we will openly share the analysis software. The long-term goal is to apply these methods to sort viruses. With this, we serve multiple research questions of the CRC VirusREvolution as well as the goals of virus description (G1, G3) and prediction of virus infectivity (G4).
- WP 1: Adaptation and optimisation of the FaNTA tool on training viruses (Schmidt)
- WP 2: Expansion of the FaNTA tool with a fluorescence readout
- WP 3: Expansion of the FaNTA tool with the iSCAT readout (Eggeling/Schmidt)
Team Members

N. N.
Doctoral Researcher

N. N.
Doctoral Researcher
Project-Specific Publications
2025
Reina, Francesco; Eggeling, Christian; Lagerholm, Christoffer
High‐Speed Interferometric Scattering Tracking Microscopy of Compartmentalized Lipid Diffusion in Living Cells Journal Article
In: ChemPhysChem, vol. 26, 2025.
@article{articlef,
title = {High‐Speed Interferometric Scattering Tracking Microscopy of Compartmentalized Lipid Diffusion in Living Cells},
author = {Francesco Reina and Christian Eggeling and Christoffer Lagerholm},
doi = {10.1002/cphc.202400407},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {ChemPhysChem},
volume = {26},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Reina, Francesco; Saavedra, Lucas; Eggeling, Christian; Barrantes, Francisco
In: Nature Communications, vol. 16, 2025.
@article{articlei,
title = {Concurrent diffusion of nicotinic acetylcholine receptors and fluorescent cholesterol disclosed by two-colour sub-millisecond MINFLUX-based single-molecule tracking},
author = {Francesco Reina and Lucas Saavedra and Christian Eggeling and Francisco Barrantes},
doi = {10.1038/s41467-025-61489-4},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Nature Communications},
volume = {16},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2024
Angelis, Giovanni; Abramo, Jacopo; Miasnikova, Mariia; Taubert, Marcel; Eggeling, Christian; Reina, Francesco
Homogeneous large field-of-view and compact iSCAT-TIRF setup for dynamic single molecule measurements Journal Article
In: Optics Express, vol. 32, pp. 46607-46620, 2024.
@article{articlee,
title = {Homogeneous large field-of-view and compact iSCAT-TIRF setup for dynamic single molecule measurements},
author = {Giovanni Angelis and Jacopo Abramo and Mariia Miasnikova and Marcel Taubert and Christian Eggeling and Francesco Reina},
doi = {10.1364/OE.532947},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {Optics Express},
volume = {32},
pages = {46607-46620},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Seltmann, Alexander; Carravilla, Pablo; Reglinski, Katharina; Eggeling, Christian; Waithe, Dominic
Neural network informed photon filtering reduces fluorescence correlation spectroscopy artifacts Journal Article
In: Biophysical Journal, vol. 123, 2024.
@article{articleg,
title = {Neural network informed photon filtering reduces fluorescence correlation spectroscopy artifacts},
author = {Alexander Seltmann and Pablo Carravilla and Katharina Reglinski and Christian Eggeling and Dominic Waithe},
doi = {10.1016/j.bpj.2024.02.012},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {Biophysical Journal},
volume = {123},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2023
Svensson, Carl-Magnus; Reglinski, Katharina; Schliebs, Wolfgang; Erdmann, Ralf; Eggeling, Christian; Figge, Marc Thilo
Quantitative analysis of peroxisome tracks using a Hidden Markov Model Journal Article
In: Sci Rep, vol. 13, no. 1, pp. 19694, 2023.
@article{Svensson:23,
title = {Quantitative analysis of peroxisome tracks using a Hidden Markov Model},
author = { Carl-Magnus Svensson and Katharina Reglinski and Wolfgang Schliebs and Ralf Erdmann and Christian Eggeling and Marc Thilo Figge},
url = {https://pubmed.ncbi.nlm.nih.gov/37951993/},
doi = {10.1038/s41598-023-46812-7},
year = {2023},
date = {2023-11-01},
urldate = {2023-11-01},
journal = {Sci Rep},
volume = {13},
number = {1},
pages = {19694},
publisher = {Springer Science and Business Media LLC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Wieduwilt, Torsten; Förster, Ronny; Nissen, Mona; Kobelke, Jens; Schmidt, Markus
Characterization of diffusing sub-10 nm nano-objects using single anti-resonant element optical fibers Journal Article
In: Nature Communications, vol. 14, 2023.
@article{articlec,
title = {Characterization of diffusing sub-10 nm nano-objects using single anti-resonant element optical fibers},
author = {Torsten Wieduwilt and Ronny Förster and Mona Nissen and Jens Kobelke and Markus Schmidt},
doi = {10.1038/s41467-023-39021-3},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {Nature Communications},
volume = {14},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2022
Nissen, Mona; Förster, Ronny; Wieduwilt, Torsten; Lorenz, Adrian; Jiang, Shiqi; Hauswald, Walter; Schmidt, Markus
Nanoparticle Tracking in Single‐Antiresonant‐Element Fiber for High‐Precision Size Distribution Analysis of Mono‐ and Polydisperse Samples Journal Article
In: Small, vol. 18, pp. 2202024, 2022.
@article{articleb,
title = {Nanoparticle Tracking in Single‐Antiresonant‐Element Fiber for High‐Precision Size Distribution Analysis of Mono‐ and Polydisperse Samples},
author = {Mona Nissen and Ronny Förster and Torsten Wieduwilt and Adrian Lorenz and Shiqi Jiang and Walter Hauswald and Markus Schmidt},
doi = {10.1002/smll.202202024},
year = {2022},
date = {2022-01-01},
urldate = {2022-01-01},
journal = {Small},
volume = {18},
pages = {2202024},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2021
Gui, Fengji; Jiang, Shiqi; Förster, Ronny; Plidschun, Malte; Weidlich, Stefan; Zhao, Jiangbo; Schmidt, Markus
Ultralong Tracking of Fast diffusing Nano‐Objects Inside Nano‐Fluidic Channel Enhanced Microstructured Optical Fiber Journal Article
In: Advanced Photonics Research, vol. 2, pp. 2100032, 2021.
@article{article,
title = {Ultralong Tracking of Fast diffusing Nano‐Objects Inside Nano‐Fluidic Channel Enhanced Microstructured Optical Fiber},
author = {Fengji Gui and Shiqi Jiang and Ronny Förster and Malte Plidschun and Stefan Weidlich and Jiangbo Zhao and Markus Schmidt},
doi = {10.1002/adpr.202100032},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Advanced Photonics Research},
volume = {2},
pages = {2100032},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2020
Förster, Ronny; Weidlich, Stefan; Nissen, Mona; Wieduwilt, Torsten; Kobelke, Jens; Goldfain, Aaron; Chiang, Timothy; Garmann, Rees; Manoharan, Vinothan; Lahini, Yoav; Schmidt, Markus
Tracking and Analyzing the Brownian Motion of Nano-objects Inside Hollow Core Fibers Journal Article
In: ACS Sensors, pp. 879–886, 2020.
@article{articled,
title = {Tracking and Analyzing the Brownian Motion of Nano-objects Inside Hollow Core Fibers},
author = {Ronny Förster and Stefan Weidlich and Mona Nissen and Torsten Wieduwilt and Jens Kobelke and Aaron Goldfain and Timothy Chiang and Rees Garmann and Vinothan Manoharan and Yoav Lahini and Markus Schmidt},
doi = {10.1021/acssensors.0c00339},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {ACS Sensors},
pages = {879–886},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2015
Faez, Sanli; Lahini, Yoav; Weidlich, Stefan; Garmann, Rees F.; Wondraczek, Katrin; Zeisberger, Matthias; Schmidt, Markus A.; Orrit, Michel; Manoharan, Vinothan N.
Fast, Label-Free Tracking of Single Viruses and Weakly Scattering Nanoparticles in a Nanofluidic Optical Fiber Journal Article
In: ACS Nano, vol. 9, no. 12, pp. 12349-12357, 2015, (PMID: 26505649).
@article{doi:10.1021/acsnano.5b05646,
title = {Fast, Label-Free Tracking of Single Viruses and Weakly Scattering Nanoparticles in a Nanofluidic Optical Fiber},
author = {Sanli Faez and Yoav Lahini and Stefan Weidlich and Rees F. Garmann and Katrin Wondraczek and Matthias Zeisberger and Markus A. Schmidt and Michel Orrit and Vinothan N. Manoharan},
url = {https://doi.org/10.1021/acsnano.5b05646},
doi = {10.1021/acsnano.5b05646},
year = {2015},
date = {2015-01-01},
urldate = {2015-01-01},
journal = {ACS Nano},
volume = {9},
number = {12},
pages = {12349-12357},
note = {PMID: 26505649},
keywords = {},
pubstate = {published},
tppubtype = {article}
}


