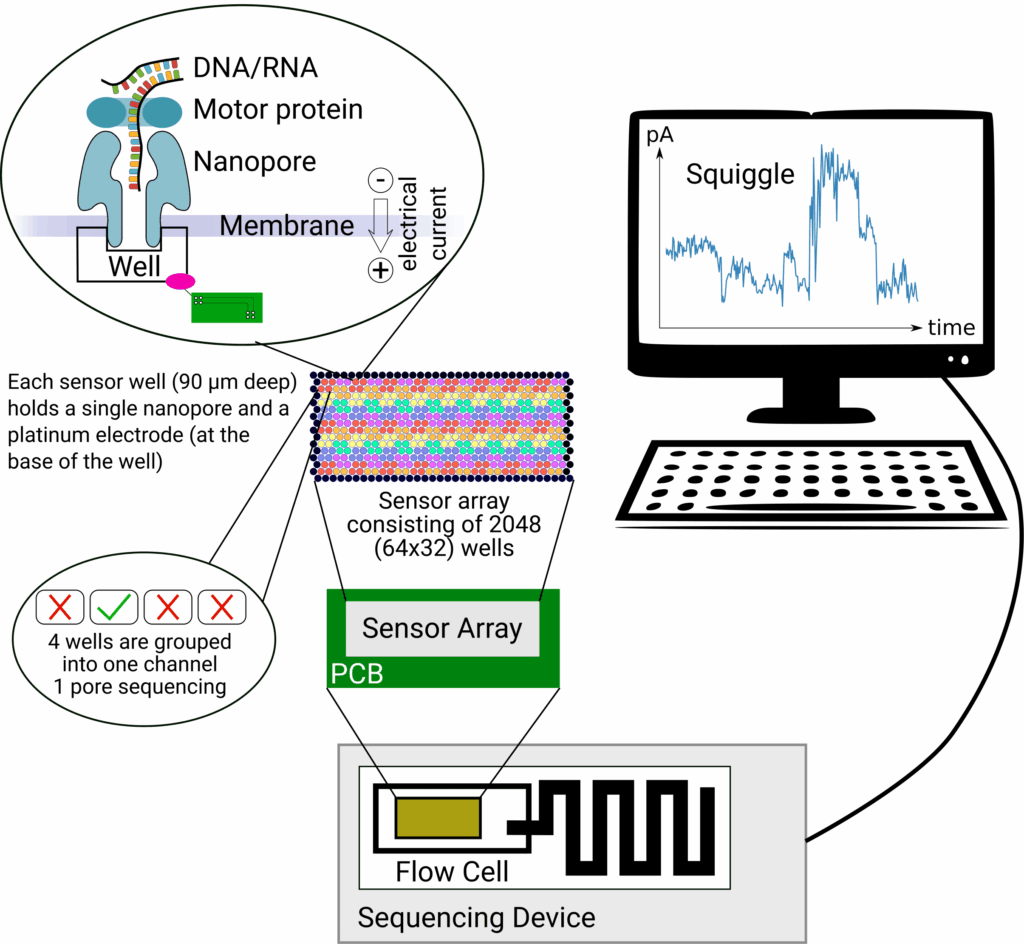
Congratulations to A04 and Z02 on their successful joint publication in Nucleic Acids Research.
Nucleic acid sequencing is the process of determining the sequence of DNA or RNA, with DNA typically used to study genomes and RNA to analyze transcriptomes. Deciphering this information has the potential to greatly advance our understanding of genomic features and cellular functions. Among the available sequencing technologies, nanopore sequencing stands out due to its unique ability to process long nucleic acid strands in real time using a compact and portable device. This enables rapid sample analysis in a wide range of laboratory and field settings. Over the past decade, nanopore sequencing has undergone rapid development and continuous refinement. Nevertheless, challenges related to protocols, data quality, and technological limitations remain. This article adopts an interdisciplinary perspective, combining experimental and computational approaches to address critical gaps in current nanopore sequencing workflows. By integrating statistically supported analyses, the study provides a comprehensive overview of the technology, from experimental design to advanced data analysis strategies. Overall, the work offers fresh perspectives and practical guidelines to help researchers overcome common challenges and maximize information gain from nanopore sequencing experiments, contributing to more robust and reproducible outcomes.