
The CRC VirusREvolution will have a unique standing in Germany and beyond. Currently, there is no other bioinformatics- and photonics-centered CRC in Germany that primarily focuses on the development of tools, particularly in the context of virus research.
CRC 1021 RNA Viruses (Marburg, 2013–2025).  1021 investigates RNA viruses, focusing on their high mutation rate induced by the lack of proofreading during replication. It focused on fundamental aspects of RNA virus replication as well as the diverse and dynamic interactions of virus factors with cellular pathways and regulatory networks that operate at the virus-host interface and determine the pathogenicity of RNA viruses in humans and animals. Although this virological CRC has ended, the comparative host cell transcriptomics analysis (nine different RNA viruses) co-coordinated by CRC VirusREvolution PI F. Weber (A03) represents a rich data source from which our CRC VirusREvolution can benefit.
1021 investigates RNA viruses, focusing on their high mutation rate induced by the lack of proofreading during replication. It focused on fundamental aspects of RNA virus replication as well as the diverse and dynamic interactions of virus factors with cellular pathways and regulatory networks that operate at the virus-host interface and determine the pathogenicity of RNA viruses in humans and animals. Although this virological CRC has ended, the comparative host cell transcriptomics analysis (nine different RNA viruses) co-coordinated by CRC VirusREvolution PI F. Weber (A03) represents a rich data source from which our CRC VirusREvolution can benefit.
 The CRC AquaDiva sought to understand the links between surface and subsurface biogeospheres. It investigated the central question of how local geology and surface conditions influence the functional diversity and ecology of the subsurface microbiome. Like other aquatic habitats, the subsurface microbiome is strongly controlled by virus activities. Under the lead of our CRC PI K. Küsel (A01), this project has generated several thousand metagenomic samples and datasets, which – although the project itself has concluded – can still greatly benefit from the application of our tools for virus discovery, annotation, and phylogenetic classification.
The CRC AquaDiva sought to understand the links between surface and subsurface biogeospheres. It investigated the central question of how local geology and surface conditions influence the functional diversity and ecology of the subsurface microbiome. Like other aquatic habitats, the subsurface microbiome is strongly controlled by virus activities. Under the lead of our CRC PI K. Küsel (A01), this project has generated several thousand metagenomic samples and datasets, which – although the project itself has concluded – can still greatly benefit from the application of our tools for virus discovery, annotation, and phylogenetic classification.  CRC 1129 studies pathogen-host interactions based on more complex models than previously used, including new technologies for the visualisation of events at high temporal and spatial resolution, quantification of microbes at the single-cell level, genetic and functional manipulation of pathogen and host factors, and establishment of novel culture systems approximating physiology.
CRC 1129 studies pathogen-host interactions based on more complex models than previously used, including new technologies for the visualisation of events at high temporal and spatial resolution, quantification of microbes at the single-cell level, genetic and functional manipulation of pathogen and host factors, and establishment of novel culture systems approximating physiology.  CRC 1278 generates polymer-based nanoparticle libraries to develop targeted anti-inflammatory strategies. The newly integrated project area D characterised particle-cell interactions. Specifically, artificial polymer-based virus-like particles are generated for targeted antivirus drug delivery. Conse- quently, our CRC VirusREvolution will support microscopy-based investigations for particle characterisation during particle-host interactions and provide virus samples for further nanoparticle production. Notably, five PIs from our CRC VirusREvolution are also involved in CRC 1278, strengthening collaboration between the two CRCs: C. Eggeling (C01, C03), V. Deckert (C02), T. Figge (C03), J. Popp (C04), and S. Höppener (Z03). We expect to use the control samples and general chemical characterisation tools and especially the open-user image facility – the Microverse Imaging Center led by our CRCs PIs C. Eggeling (C01, C03) and J. Popp (C04)– which is also supported by the CRC 1278 and will be available to all researchers from the CRC VirusREvolution.
CRC 1278 generates polymer-based nanoparticle libraries to develop targeted anti-inflammatory strategies. The newly integrated project area D characterised particle-cell interactions. Specifically, artificial polymer-based virus-like particles are generated for targeted antivirus drug delivery. Conse- quently, our CRC VirusREvolution will support microscopy-based investigations for particle characterisation during particle-host interactions and provide virus samples for further nanoparticle production. Notably, five PIs from our CRC VirusREvolution are also involved in CRC 1278, strengthening collaboration between the two CRCs: C. Eggeling (C01, C03), V. Deckert (C02), T. Figge (C03), J. Popp (C04), and S. Höppener (Z03). We expect to use the control samples and general chemical characterisation tools and especially the open-user image facility – the Microverse Imaging Center led by our CRCs PIs C. Eggeling (C01, C03) and J. Popp (C04)– which is also supported by the CRC 1278 and will be available to all researchers from the CRC VirusREvolution. 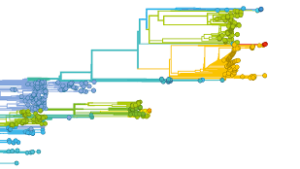 CRC 1310 investigates whether and how future evolutionary pathways and outcomes can be predicted by studying rapidly evolving systems such as microbial populations, viruses and immune repertoires, and cancer-cell populations. The centre develops predictive approaches for drug-resistance evolution, vaccine design, and immune escape, but no virulence. CRC 1310 is therefore highly complementary to our CRC VirusREvolution, as the evolutionary insights and predictive models can directly benefit virus-host interaction studies and computational tool development.
CRC 1310 investigates whether and how future evolutionary pathways and outcomes can be predicted by studying rapidly evolving systems such as microbial populations, viruses and immune repertoires, and cancer-cell populations. The centre develops predictive approaches for drug-resistance evolution, vaccine design, and immune escape, but no virulence. CRC 1310 is therefore highly complementary to our CRC VirusREvolution, as the evolutionary insights and predictive models can directly benefit virus-host interaction studies and computational tool development.  (Giessen, since 2018). The RTG 2355 addresses RNA-based regulatory mechanisms in a broad range of model organisms, including viruses. It brings together experts with different backgrounds, e.g., biochemists, bioinformaticians, and virologists like RTG 2355 member F. Weber (A03). The massive concentration of expertise on all aspects of RNA biology makes RTG 2355 highly synergistic with our CRC VirusREvolution.
(Giessen, since 2018). The RTG 2355 addresses RNA-based regulatory mechanisms in a broad range of model organisms, including viruses. It brings together experts with different backgrounds, e.g., biochemists, bioinformaticians, and virologists like RTG 2355 member F. Weber (A03). The massive concentration of expertise on all aspects of RNA biology makes RTG 2355 highly synergistic with our CRC VirusREvolution.GRK 2485 VIPER (Hannover, since 2019). This Research Training Group focuses on virus detection, pathogenesis, and intervention in animals. It aims to train the next-generation scientists in virus emergence (discovery, isolation, characterisation, surveillance, and pathogenesis). These are mainly experimental projects. Deputy Speaker Paul Becher welcomes the development of new tools in all of these areas to enhance the research of GK 2485.
This Research Training Group focuses on virus detection, pathogenesis, and intervention in animals. It aims to train the next-generation scientists in virus emergence (discovery, isolation, characterisation, surveillance, and pathogenesis). These are mainly experimental projects. Deputy Speaker Paul Becher welcomes the development of new tools in all of these areas to enhance the research of GK 2485.
 This Cluster of Excellence, led by K. Küsel (A01), addresses questions like: (i) What defines the dynamics of microbial balance? (ii) Which factors disturb the equilibrium, and how is re-balancing attained? (iii) How can we shape and control microbial consortia to create a beneficial impact? These questions are addressed at multiple levels, and the CRC VirusREvolution perfectly complements EXC 2051 by introducing viruses – key regulators of microbial communities – into the research focus. The envisioned collaboration focuses on our development of tools that the Cluster of Excellence will employ to advance knowledge of the roles viruses play in interactions with other organisms. Notably, four further PIs from our CRC VirusREvolution are also main PIs of EXC 2051, strengthening collaboration between the two initiatives: B. Dutilh (A01), M. Marz (A04), C. Eggeling (C01, C03), and J. Popp (C04). Additionally, C. Eggeling (C01, C03) and J. Popp (C04) are leading the open-user image facility, the Microverse Imaging Center, which will also be available to all researchers from the CRC VirusREvolution.
This Cluster of Excellence, led by K. Küsel (A01), addresses questions like: (i) What defines the dynamics of microbial balance? (ii) Which factors disturb the equilibrium, and how is re-balancing attained? (iii) How can we shape and control microbial consortia to create a beneficial impact? These questions are addressed at multiple levels, and the CRC VirusREvolution perfectly complements EXC 2051 by introducing viruses – key regulators of microbial communities – into the research focus. The envisioned collaboration focuses on our development of tools that the Cluster of Excellence will employ to advance knowledge of the roles viruses play in interactions with other organisms. Notably, four further PIs from our CRC VirusREvolution are also main PIs of EXC 2051, strengthening collaboration between the two initiatives: B. Dutilh (A01), M. Marz (A04), C. Eggeling (C01, C03), and J. Popp (C04). Additionally, C. Eggeling (C01, C03) and J. Popp (C04) are leading the open-user image facility, the Microverse Imaging Center, which will also be available to all researchers from the CRC VirusREvolution. (Jena, 2020–2025). The Marie Skłodowska-Curie International Training Network VIROINF, which concluded recently, successfully fostered cross-disciplinary collaboration between virology and bioinformatics through its tandem project concept, pairing wet lab and computational tool development. Led by M. Marz (A04), VIROINF’s experience has been invaluable. Building on this strong foundation, CRC VirusREvolution will integrate the combined knowledge – particularly in virus-host interactions – during its later funding phases. Tools developed within VIROINF will be integrated into the planned overall platform of CRC VirusREvolution during its second and third funding periods.
(Jena, 2020–2025). The Marie Skłodowska-Curie International Training Network VIROINF, which concluded recently, successfully fostered cross-disciplinary collaboration between virology and bioinformatics through its tandem project concept, pairing wet lab and computational tool development. Led by M. Marz (A04), VIROINF’s experience has been invaluable. Building on this strong foundation, CRC VirusREvolution will integrate the combined knowledge – particularly in virus-host interactions – during its later funding phases. Tools developed within VIROINF will be integrated into the planned overall platform of CRC VirusREvolution during its second and third funding periods.  (since 2020). PREVIR is an interdisciplinary initiative that integrates genetic, epidemiological, and antigenic data to predict the evolutionary trajectories of viruses, including Influenza A virus and SARS-CoV-2. It develops computational frameworks to forecast variant emergence, assess mutation-driven fitness changes, and identify lineage dynamics relevant for public health. PREVIR’s predictive modelling capabilities align extremely well with the scientific goals of CRC VirusREvolution. They will provide complementary data streams and analytical approaches that enhance our ability to interpret and anticipate viral evolution.
(since 2020). PREVIR is an interdisciplinary initiative that integrates genetic, epidemiological, and antigenic data to predict the evolutionary trajectories of viruses, including Influenza A virus and SARS-CoV-2. It develops computational frameworks to forecast variant emergence, assess mutation-driven fitness changes, and identify lineage dynamics relevant for public health. PREVIR’s predictive modelling capabilities align extremely well with the scientific goals of CRC VirusREvolution. They will provide complementary data streams and analytical approaches that enhance our ability to interpret and anticipate viral evolution.  (since 2021). This research group investigates the early events of nuclear DNA virus infection. It aims to open up new possibilities to understand and ultimately control acute and chronic DNA virus infections. The development of bioinformatic tools will largely focus on integrative analysis of multiomics data, such as single-cell RNA-seq and scSLAM-seq. C. Friedel (A02) is involved in the functional analysis. We do not see an overlap with the CRC VirusREvolution.
(since 2021). This research group investigates the early events of nuclear DNA virus infection. It aims to open up new possibilities to understand and ultimately control acute and chronic DNA virus infections. The development of bioinformatic tools will largely focus on integrative analysis of multiomics data, such as single-cell RNA-seq and scSLAM-seq. C. Friedel (A02) is involved in the functional analysis. We do not see an overlap with the CRC VirusREvolution.  (since 2021). The single projects of SPP 2330 use intensive omics techniques to analyse phage-host interactions. These aim to discover fundamentally new concepts and mechanisms in virus organisation, unicellular and multicellular antiviral defences, and virus impacts on microbial communities. Notably, B. Dutilh (A01) and K. Papenfort (A04) are leading a project within SPP 2330. Apart from the Z-project, which generates a database for phage-induced data, no SPP 2330 project develops tools for specifically analysing virus data. The tools developed in the CRC VirusREvolution are made directly available to member of SPP 2330.
(since 2021). The single projects of SPP 2330 use intensive omics techniques to analyse phage-host interactions. These aim to discover fundamentally new concepts and mechanisms in virus organisation, unicellular and multicellular antiviral defences, and virus impacts on microbial communities. Notably, B. Dutilh (A01) and K. Papenfort (A04) are leading a project within SPP 2330. Apart from the Z-project, which generates a database for phage-induced data, no SPP 2330 project develops tools for specifically analysing virus data. The tools developed in the CRC VirusREvolution are made directly available to member of SPP 2330. SARS-CoV-2Dx (Jena, since 2021).  The BMFTR-funded joint project “SARS-CoV-2Dx” brings together leading partners from Friedrich Schiller University Jena (FSU), Jena University Hospital, the Leibniz Institute of Photonic Technology, and the Leibniz Hans Knoell Institute for Natural Product Research and Infection Biology. The consortium, coordinated by S. Deinhardt-Emmer (C02), includes several PIs active within our CRC, such as V. Deckert (C02) and J. Popp (C04). The project aims to develop innovative diagnostic and therapeutic strategies for virus infections, combining advanced photonic technologies with infection biology to improve the rapid detection and targeted treatment of pathogens such as SARS-CoV-2.
The BMFTR-funded joint project “SARS-CoV-2Dx” brings together leading partners from Friedrich Schiller University Jena (FSU), Jena University Hospital, the Leibniz Institute of Photonic Technology, and the Leibniz Hans Knoell Institute for Natural Product Research and Infection Biology. The consortium, coordinated by S. Deinhardt-Emmer (C02), includes several PIs active within our CRC, such as V. Deckert (C02) and J. Popp (C04). The project aims to develop innovative diagnostic and therapeutic strategies for virus infections, combining advanced photonic technologies with infection biology to improve the rapid detection and targeted treatment of pathogens such as SARS-CoV-2.
NFDI4Microbiota  (Cologne, since 2021). This national research infrastructure profits greatly from the CRC VirusREvolution by the improvement of the virus database VirJenDB. Under the leadership of N. Cassman (Z02), the NFDI4Microbiota team supports the acquisition of virus sequences and annotations, manages storage of datasets in VirJenDB and other databases, and coordinates data workflows with relevant NFDI consortia. The doctoral researcher funded from CRC VirusREvolution, also supervised by N. Cassman (Z02), will carry out three tasks (WP 3 of Z02) that are distinct from the NFDI4Microbiota agenda: (i) Integration of CRC VirusREvolution tools into VirJenDB via CLOWM; (ii) Data integration and harmonisation of heterogeneous datasets to prepare for novel knowledge; (iii) Development of comprehensive visualisation features for the integrated data and analyses.
(Cologne, since 2021). This national research infrastructure profits greatly from the CRC VirusREvolution by the improvement of the virus database VirJenDB. Under the leadership of N. Cassman (Z02), the NFDI4Microbiota team supports the acquisition of virus sequences and annotations, manages storage of datasets in VirJenDB and other databases, and coordinates data workflows with relevant NFDI consortia. The doctoral researcher funded from CRC VirusREvolution, also supervised by N. Cassman (Z02), will carry out three tasks (WP 3 of Z02) that are distinct from the NFDI4Microbiota agenda: (i) Integration of CRC VirusREvolution tools into VirJenDB via CLOWM; (ii) Data integration and harmonisation of heterogeneous datasets to prepare for novel knowledge; (iii) Development of comprehensive visualisation features for the integrated data and analyses.
 (Jena, since 2021). The Leibniz Center for Photonics in Infection Research (LPI) will establish a user-friendly one-stop infrastructure for photonic and optical technologies in infection research. Fundamentally new solutions for the diagnosis, monitoring, and therapy of infections will be researched and developed in order to transfer these solutions rapidly into routine applications by 2027. Scientists within the LPI, of which many are also PIs of the CRC VirusREvolution, are currently working on basic technologies in five collaborative projects (multimodal imaging techniques, light-based technologies, evaluation methods of photonic data, modelling immune responses, and new therapy concepts), which will form the unique technological infrastructure of LPI alongside state-of-the-art technologies.
(Jena, since 2021). The Leibniz Center for Photonics in Infection Research (LPI) will establish a user-friendly one-stop infrastructure for photonic and optical technologies in infection research. Fundamentally new solutions for the diagnosis, monitoring, and therapy of infections will be researched and developed in order to transfer these solutions rapidly into routine applications by 2027. Scientists within the LPI, of which many are also PIs of the CRC VirusREvolution, are currently working on basic technologies in five collaborative projects (multimodal imaging techniques, light-based technologies, evaluation methods of photonic data, modelling immune responses, and new therapy concepts), which will form the unique technological infrastructure of LPI alongside state-of-the-art technologies.  (Hamburg, since 2024). CRC 1648 focuses on understanding the molecular mechanisms that regulate cellular metabolism and their implications for health and disease. The collaborative research centre investigates metabolic pathways, signalling networks, and their interplay in various biological contexts. By combining experimental and computational approaches, the projects aim to uncover how metabolic dysregulation contributes to conditions such as cancer, diabetes, and neurodegeneration. The insights gained are expected to inform the development of novel therapeutic strategies targeting metabolic processes. CRC 1648 is a perfect complement to our CRC VirusREvolution, as they focus on applying computational tools, while we specialise in developing them. With both initiatives working around the same time, a strong and synergistic interaction is planned to maximise scientific impact.
(Hamburg, since 2024). CRC 1648 focuses on understanding the molecular mechanisms that regulate cellular metabolism and their implications for health and disease. The collaborative research centre investigates metabolic pathways, signalling networks, and their interplay in various biological contexts. By combining experimental and computational approaches, the projects aim to uncover how metabolic dysregulation contributes to conditions such as cancer, diabetes, and neurodegeneration. The insights gained are expected to inform the development of novel therapeutic strategies targeting metabolic processes. CRC 1648 is a perfect complement to our CRC VirusREvolution, as they focus on applying computational tools, while we specialise in developing them. With both initiatives working around the same time, a strong and synergistic interaction is planned to maximise scientific impact. (Jena, from 2026). This German Research Training Group will study how infectious diseases (especially those caused by viruses) lead to neurological or psychiatric after-effects following infection. The group is interested not only in the clinical symptoms but also in mechanisms, including how viruses act or trigger processes that affect the brain and mind. Our CRC PI M. Marz (A04) is co-speaker of this RTG, which leads to a close collaboration between us for a direct application of our tools in clinics and to possibly connect virus variants to psychiatric symptoms in the future.
(Jena, from 2026). This German Research Training Group will study how infectious diseases (especially those caused by viruses) lead to neurological or psychiatric after-effects following infection. The group is interested not only in the clinical symptoms but also in mechanisms, including how viruses act or trigger processes that affect the brain and mind. Our CRC PI M. Marz (A04) is co-speaker of this RTG, which leads to a close collaboration between us for a direct application of our tools in clinics and to possibly connect virus variants to psychiatric symptoms in the future.