
A01: Do the shape and size of quasispecies reflect the host range of viruses?

Projects of the CRC 1768

A01: Do the shape and size of quasispecies reflect the host range of viruses?
Viruses exist as dynamic populations of closely related viral genomes arising from mutations, known as quasispecies. We hypothesise that viruses use their quasispecies to expand their evolutionary potential, making them critical for adaptation to new hosts and for resistance to host defences or immunity. Yet, the evolutionary trajectories of viruses cannot be fully understood without considering their ecological context. Host range and environmental conditions act as powerful filters and drivers of viral diversification, raising fundamental research questions at the interface of viral ecology and evolution: How do host interactions and environmental factors shape the emergence, stability, and adaptability of viral quasispecies? If the genetic diversity of the quasispecies reflects the evolutionary potential and ecological interactions of the virus, by which molecular mechanisms do viruses exploit their quasispecies for host range evolution? To address these fundamental questions at the core of our project, we will develop and apply a novel suite of computational tools based on Sequence Variation Graphs (SVGs). SVGs are increasingly utilised for population structure analysis in higher organisms, but their application in virology is limited due to the high mutation rates and genomic diversity of viruses. Nevertheless, they offer potential for analysing data from both genomic and metagenomic samples. In work package WP 1, we will build a quasispecies Sequence Variation Graph (qs-SVG) toolkit that can store sequencing data of viral populations, and we will use and further improve the tool in the remaining work packages. Once the tool is built, we will first apply it to an ideal case where abundant data is available, i.e., SARS-CoV-2 and influenza viruses before, taking on a more challenging case, i.e., bacteriophages found in environmental metagenomes.
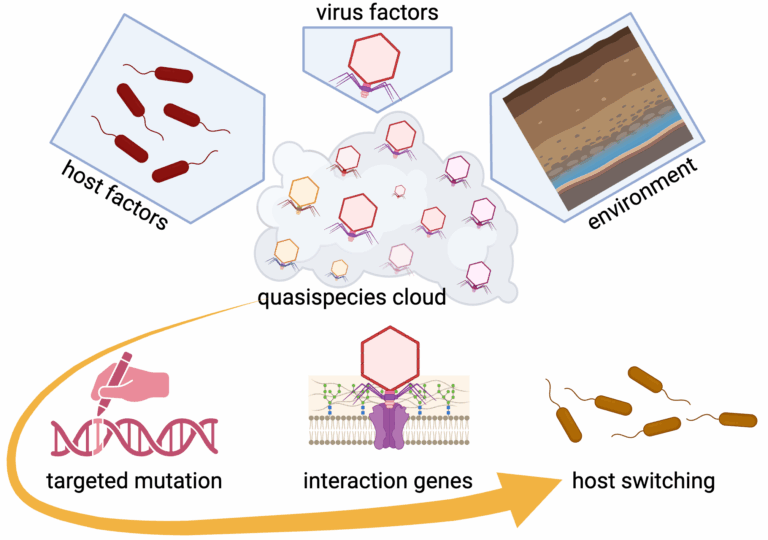
The central hypothesis of our project is that viruses use their quasispecies for host range evolution, with specific hypotheses including: (1) Viral quasispecies are shaped by host- and virus-specific factors, and by environmental context; (2) Quasispecies are shaped by targeted mutations, specifically of host interaction genes, leading to host switching.
By combining these two study systems in our project, we will be able to test different functionalities of the qs-SVG toolkit and develop an optimal bioinformatic solution. In WP 2, we will exploit new and existing data on the quasispecies of human viruses and bacteriophages with broad and narrow host ranges, to test the specific hypothesis that viruses with a broad host range also have a large quasispecies. In WP 3, we will investigate under what conditions viruses evolve their quasispecies. We will examine the relationship between quasispecies, host diversity, and the environment, using both in vitro data from isolates, and in situ data by screening metagenomic data sets derived from environmental samples. Finally, in WP 4 we will focus on the underlying mutational mechanisms. How does quasispecies sequence variation arise? Can viruses exploit mutation to expand their host range, and what molecular mechanisms enable this?
The qs-SVG toolkit will provide immediate access to mutations and genetic functions, enabling us to explore the association of these features with variable sites. We will also chart the distribution of the mutational mechanisms across ecosystems and host types (G3). Thus, our new tool suite will facilitate describing, quantifying, and understanding emerging viruses (G1) and offer new perspectives for studying their evolution (G2) in the context of host range.
Project Overview
Based on our preliminary results, viral quasispecies may be accessed by exploiting raw sequencing data, whether from isolates or from enriched complex samples, but existing read-mapping approaches cannot reveal all relevant mutations, as explained above. Thus, we need new analysis tools that are capable of tracking different types of mutations that occur in naturally evolving viral populations. We will develop a bioinformatic tool suite based on SVGs to quantify the mutations in an evolving viral population, extract the trajectories of sequence features (e.g. specific alleles) across datasets (time series), and statistically assess their association with metadata, such as host-range measurements. Furthermore, we will use existing and newly generated data to investigate the ecological imprint on quasispecies, including whether the mutational profile can predict the host range of viruses in eukaryotic and microbial systems. By combining host range profiling, in situ virus-host mapping, and evolutionary co-culture experiments, we aim to uncover how viral microdiversity enables host switching and ecological flexibility. We will test our hypothesis that large quasispecies provide a mutational reservoir that increases potential host range – especially in complex environments like soil – while small quasispecies reflect a narrower host range in stable, low-diversity systems such as groundwater. Finally, we will investigate to what extent these patterns are virus- and host-specific, and associated with mutations that affect specific functional categories of genes. This work bridges viral population genetics with ecological theory to better understand the evolutionary dynamics of virus–host interactions.
- Tool to be developed: A software suite, the quasispecies Sequence Variation Graph (qs-SVG) toolkit, to capture quantify, and investigate viral quasispecies. The qs-SVG toolkit will store diverse sequencing data from viral populations based on complete genomes and unassembled sequencing data, facilitate access to mutations, mutational mechanisms, functional annotations in viral genome sequences, and allow their quantification.
Hypothesis enabled by the proposed tool: The following central and specific hypotheses guide this project.
- Viral quasispecies are shaped by virus- and host-specific factors, and by environmental context
- Host factors: New or suboptimal hosts, recent host switches, diverse hosts, or hosts with a wide array of defence strategies will cause viruses to expand their quasispecies.
- Virus factors: Viruses with rapid genome evolution have large quasispecies, including those encoding diversity-generating elements such as error-prone polymerases or reverse transcriptase-mediated targeted editing, or temperate bacteriophages with high recombination rates.
- Environment: Viruses have large quasispecies in complex environments, e.g., in soil, where the potential for encountering different host cells is high, compared to more homogeneous systems such as groundwater, where hosts are genotypically and phenotypically similar.
- Quasispecies can reveal novel information about viral functions and phenotypes.
- Quasispecies are shaped by targeted evolvability mechanisms such as diversity-generating elements. These molecular mechanisms may be revealed by analysing mutational patterns and may be biotechnologically or clinically relevant.
- Mutations differentially affect specific functional categories of genes, such as receptor-binding proteins and anti-defense genes. Charting quasispecies mutations will thus reveal critical new information about genes involved in host interaction, contributing new insights into tropism and zoonosis.
- Viruses exploit their quasispecies to adapt to different hosts. Viruses with large quasispecies have a broader host range and/or more readily switch to a new host than viruses with small quasispecies.
Overarching CRC goals: Our project develops and applies a quasispecies Sequence Variation Graph (qs-SVG) toolkit to capture, annotate, and quantify intra-population viral diversity from long/short-read and metagenomic data, enabling rapid characterization of emerging viruses and their mutational mechanisms (G1). By deploying qs-SVG across human viruses and environmental phages, the project dissects how ecological context and host diversity shape quasispecies structure, deriving generalisable rules and experimentally testable trajectories of host-range evolution (G2, G3).
Work Packages (WP):
- WP 1: Development of a tool to formally quantify quasispecies (Dutilh)
- WP 2: Measuring the effect of quasispecies on host range and other phenotypic characters (Dutilh/Küsel)
- WP 3: Measuring quasispecies in natural ecosystems and targeted microcosm incubations (Küsel/Dutilh)
- WP 4: Ecological imprint of evolvability mechanisms and implications for classification (Dutilh/Küsel)
Team Members
2025
Zielezinski, Andrzej; Gudyś, Adam; Barylski, Jakub; Siminski, Krzysztof; Rozwalak, Piotr; Dutilh, Bas E; Deorowicz, Sebastian
Ultrafast and accurate sequence alignment and clustering of viral genomes. Journal Article
In: Nat Methods, vol. 22, iss. 6, pp. 1191–1194, 2025, ISSN: 1548-7105.
@article{Zielezinski:25,
title = {Ultrafast and accurate sequence alignment and clustering of viral genomes.},
author = {Andrzej Zielezinski and Adam Gudyś and Jakub Barylski and Krzysztof Siminski and Piotr Rozwalak and Bas E Dutilh and Sebastian Deorowicz},
url = {https://pubmed.ncbi.nlm.nih.gov/40374946/},
doi = {10.1038/s41592-025-02701-7},
issn = {1548-7105},
year = {2025},
date = {2025-06-01},
journal = {Nat Methods},
volume = {22},
issue = {6},
pages = {1191–1194},
keywords = {},
pubstate = {ppublish},
tppubtype = {article}
}
Pratama, Akbar Adjie; Perez-Carrascal, Olga; Sullivan, Matthew B.; Küsel, Kirsten
Hidden viral players: Diversity and ecological roles of viruses in groundwater microbiomes Journal Article
In: 2025.
@article{Pratama:25,
title = {Hidden viral players: Diversity and ecological roles of viruses in groundwater microbiomes},
author = {Akbar Adjie Pratama and Olga Perez-Carrascal and Matthew B. Sullivan and Kirsten Küsel},
doi = {10.1101/2025.05.30.656956},
year = {2025},
date = {2025-05-01},
publisher = {Cold Spring Harbor Laboratory},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2024
Beeloo, Rick; Zomer, Aldert L; Deorowicz, Sebastian; Dutilh, Bas E
Graphite: Painting genomes using a colored de Bruijn graph Journal Article
In: NAR Genom Bioinform, vol. 6, no. 4, 2024, ISSN: 2631-9268.
@article{Beeloo:24,
title = {Graphite: Painting genomes using a colored de Bruijn graph},
author = {Rick Beeloo and Aldert L Zomer and Sebastian Deorowicz and Bas E Dutilh},
doi = {10.1093/nargab/lqae142},
issn = {2631-9268},
year = {2024},
date = {2024-09-01},
journal = {NAR Genom Bioinform},
volume = {6},
number = {4},
publisher = {Oxford University Press (OUP)},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Chaudhari, Narendrakumar M.; Pérez-Carrascal, Olga M.; Overholt, Will A.; Totsche, Kai U.; Küsel, Kirsten
Genome streamlining in Parcubacteria transitioning from soil to groundwater Journal Article
In: Environ Microbiome, vol. 19, no. 1, 2024, ISSN: 2524-6372.
@article{Chaudhari:24,
title = {Genome streamlining in Parcubacteria transitioning from soil to groundwater},
author = {Narendrakumar M. Chaudhari and Olga M. Pérez-Carrascal and Will A. Overholt and Kai U. Totsche and Kirsten Küsel},
doi = {10.1186/s40793-024-00581-6},
issn = {2524-6372},
year = {2024},
date = {2024-06-01},
journal = {Environ Microbiome},
volume = {19},
number = {1},
publisher = {Springer Science and Business Media LLC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2023
Meijenfeldt, F. A. Bastiaan; Hogeweg, Paulien; Dutilh, Bas E.
A social niche breadth score reveals niche range strategies of generalists and specialists Journal Article
In: Nat Ecol Evol, vol. 7, no. 5, pp. 768–781, 2023.
@article{Meijenfeldt:23,
title = {A social niche breadth score reveals niche range strategies of generalists and specialists},
author = {F. A. Bastiaan Meijenfeldt and Paulien Hogeweg and Bas E. Dutilh},
doi = {10.1038/s41559-023-02027-7},
year = {2023},
date = {2023-01-01},
journal = {Nat Ecol Evol},
volume = {7},
number = {5},
pages = {768–781},
publisher = {Springer Science and Business Media LLC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2022
Overholt, Will A.; Trumbore, Susan; Xu, Xiaomei; Bornemann, Till L. V.; Probst, Alexander J.; Krüger, Markus; Herrmann, Martina; Thamdrup, Bo; Bristow, Laura A.; Taubert, Martin; Schwab, Valérie F.; Hölzer, Martin; Marz, Manja; Küsel, Kirsten
Carbon fixation rates in groundwater similar to those in oligotrophic marine systems Journal Article
In: Nat Geosci, vol. 15, no. 7, pp. 561–567, 2022, ISSN: 1752-0908.
@article{Overholt:22,
title = {Carbon fixation rates in groundwater similar to those in oligotrophic marine systems},
author = {Will A. Overholt and Susan Trumbore and Xiaomei Xu and Till L. V. Bornemann and Alexander J. Probst and Markus Krüger and Martina Herrmann and Bo Thamdrup and Laura A. Bristow and Martin Taubert and Valérie F. Schwab and Martin Hölzer and Manja Marz and Kirsten Küsel},
doi = {10.1038/s41561-022-00968-5},
issn = {1752-0908},
year = {2022},
date = {2022-06-01},
journal = {Nat Geosci},
volume = {15},
number = {7},
pages = {561–567},
publisher = {Springer Science and Business Media LLC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Meaden, Sean; Biswas, Ambarish; Arkhipova, Ksenia; Morales, Sergio E.; Dutilh, Bas E.; Westra, Edze R.; Fineran, Peter C.
High viral abundance and low diversity are associated with increased CRISPR-Cas prevalence across microbial ecosystems Journal Article
In: Curr Biol, vol. 32, iss. 1, pp. 220–227.e5, 2022.
@article{Meaden:22,
title = {High viral abundance and low diversity are associated with increased CRISPR-Cas prevalence across microbial ecosystems},
author = {Sean Meaden and Ambarish Biswas and Ksenia Arkhipova and Sergio E. Morales and Bas E. Dutilh and Edze R. Westra and Peter C. Fineran},
doi = {10.1016/j.cub.2021.10.038},
year = {2022},
date = {2022-01-01},
journal = {Curr Biol},
volume = {32},
issue = {1},
pages = {220–227.e5},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2019
Gregory, Ann C.; Zayed, Ahmed A.; Conceição-Neto, Nádia; Temperton, Ben; Bolduc, Ben; Alberti, Adriana; Ardyna, Mathieu; Arkhipova, Ksenia; Carmichael, Margaux; Cruaud, Corinne; Dimier, Céline; Domínguez-Huerta, Guillermo; Ferland, Joannie; Kandels, Stefanie; Liu, Yunxiao; Marec, Claudie; Pesant, Stéphane; Picheral, Marc; Pisarev, Sergey; Poulain, Julie; Tremblay, Jean-Éric; Vik, Dean; Babin, Marcel; Bowler, Chris; Culley, Alexander I.; Vargas, Colomban; Dutilh, Bas E.; Iudicone, Daniele; Karp-Boss, Lee; Roux, Simon; Sunagawa, Shinichi; Wincker, Patrick; Sullivan, Matthew B.; Acinas, Silvia G.; Babin, Marcel; Bork, Peer; Boss, Emmanuel; Bowler, Chris; Cochrane, Guy; Vargas, Colomban; Follows, Michael; Gorsky, Gabriel; Grimsley, Nigel; Guidi, Lionel; Hingamp, Pascal; Iudicone, Daniele; Jaillon, Olivier; Kandels-Lewis, Stefanie; Karp-Boss, Lee; Karsenti, Eric; Not, Fabrice; Ogata, Hiroyuki; Pesant, Stéphane; Poulton, Nicole; Raes, Jeroen; Sardet, Christian; Speich, Sabrina; Stemmann, Lars; Sullivan, Matthew B.; Sunagawa, Shinichi; Wincker, Patrick
Marine DNA viral macro- and microdiversity from pole to pole Journal Article
In: Cell, vol. 177, iss. 5, no. 5, pp. 1109–1123.e14, 2019, ISSN: 0092-8674.
@article{Gregory:19,
title = {Marine DNA viral macro- and microdiversity from pole to pole},
author = {Ann C. Gregory and Ahmed A. Zayed and Nádia Conceição-Neto and Ben Temperton and Ben Bolduc and Adriana Alberti and Mathieu Ardyna and Ksenia Arkhipova and Margaux Carmichael and Corinne Cruaud and Céline Dimier and Guillermo Domínguez-Huerta and Joannie Ferland and Stefanie Kandels and Yunxiao Liu and Claudie Marec and Stéphane Pesant and Marc Picheral and Sergey Pisarev and Julie Poulain and Jean-Éric Tremblay and Dean Vik and Marcel Babin and Chris Bowler and Alexander I. Culley and Colomban Vargas and Bas E. Dutilh and Daniele Iudicone and Lee Karp-Boss and Simon Roux and Shinichi Sunagawa and Patrick Wincker and Matthew B. Sullivan and Silvia G. Acinas and Marcel Babin and Peer Bork and Emmanuel Boss and Chris Bowler and Guy Cochrane and Colomban Vargas and Michael Follows and Gabriel Gorsky and Nigel Grimsley and Lionel Guidi and Pascal Hingamp and Daniele Iudicone and Olivier Jaillon and Stefanie Kandels-Lewis and Lee Karp-Boss and Eric Karsenti and Fabrice Not and Hiroyuki Ogata and Stéphane Pesant and Nicole Poulton and Jeroen Raes and Christian Sardet and Sabrina Speich and Lars Stemmann and Matthew B. Sullivan and Shinichi Sunagawa and Patrick Wincker},
doi = {10.1016/j.cell.2019.03.040},
issn = {0092-8674},
year = {2019},
date = {2019-05-01},
journal = {Cell},
volume = {177},
number = {5},
issue = {5},
pages = {1109–1123.e14},
publisher = {Elsevier BV},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Jonge, Patrick A; Nobrega, Franklin L; Brouns, Stan J J; Dutilh, Bas E
Molecular and evolutionary determinants of bacteriophage host range Journal Article
In: Trends Microbiol, vol. 27, iss. 1, pp. 51–63, 2019.
@article{deJonge:19,
title = {Molecular and evolutionary determinants of bacteriophage host range},
author = {Patrick A Jonge and Franklin L Nobrega and Stan J J Brouns and Bas E Dutilh},
doi = {10.1016/j.tim.2018.08.006},
year = {2019},
date = {2019-01-01},
journal = {Trends Microbiol},
volume = {27},
issue = {1},
pages = {51–63},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2018
Geesink, Patricia; Tyc, Olaf; Küsel, Kirsten; Taubert, Martin; Velde, Charlotte; Kumar, Swatantar; Garbeva, Paolina
Growth promotion and inhibition induced by interactions of groundwater bacteria. Journal Article
In: FEMS Microbiol Ecol, vol. 94, iss. 11, 2018, ISSN: 1574-6941.
@article{Geesink:18,
title = {Growth promotion and inhibition induced by interactions of groundwater bacteria.},
author = {Patricia Geesink and Olaf Tyc and Kirsten Küsel and Martin Taubert and Charlotte Velde and Swatantar Kumar and Paolina Garbeva},
url = {https://pubmed.ncbi.nlm.nih.gov/30137328/},
doi = {10.1093/femsec/fiy164},
issn = {1574-6941},
year = {2018},
date = {2018-11-01},
journal = {FEMS Microbiol Ecol},
volume = {94},
issue = {11},
abstract = {Microorganisms can produce a plethora of secondary metabolites, some acting as signaling compounds and others as suppressing agents. As yet, the potential of groundwater microbes to produce antimicrobial compounds to increase their competitiveness against other bacteria has not been examined. In this study, we developed an AlamarBlue® based high-throughput screening method that allowed for a fast and highly standardized evaluation of both growth-inhibiting and -promoting metabolites. With this technique, 149 screened bacterial isolates were grown in monocultures and in 1402 co-cultures. Co-cultivation did not increase the frequency of growth inhibition against the two tested model organisms (Staphylococcus aureus 533R4 and Escherichia coli WA321) compared to monocultures. Mainly co-cultivation of Proteobacteria induced growth inhibition of both model organisms. Only slightly increased growth promotion of S. aureus 533R4 was observed. Growth-promoting effects on E. coli WA321 were observed by supernatants from co-cultures between Bacteroidetes and Firmicutes. With the standardized screening for both growth-inhibiting and -promoting effects, this method will enable further studies to elaborate and better understand complex inter-specific interactions and networks in aquatic communities as well as in other environments.},
keywords = {},
pubstate = {ppublish},
tppubtype = {article}
}




