
C03: Deciphering virus-membrane interactions with advanced optical microscopy and machine

Projects of the CRC 1768

C03: Deciphering virus-membrane interactions with advanced optical microscopy and machine
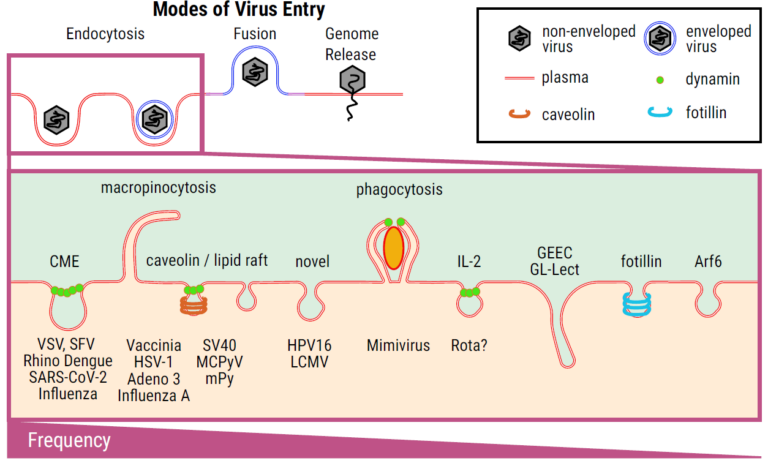
Sketch of different uptake mechanisms, ranging from various endocytosis pathways to membrane fusion to the injection of genetic material (as for bacteriophages).
Finally, data analysis needs to be tailored to correlate a “microscopic signature” of entry events with fusion signatures and particular endocytic mechanisms, which would then make it possible to identify the mode(s) of entry of an emerging virus. Unfortunately, the realisation of high-resolution microscope experiments of virus uptake is usually too complex to accomplish a straightforward and quick prediction of entry modes of newly emerging viruses. This is, however, within the overall aim of this CRC VirusREvolution, which is to set up tools to quickly react to potential pandemic threats. We therefore propose to realise a fast and facile tool to predict the mode(s) of virus entry from standard low-resolution live-cell microscopy data. To achieve this, we will combine high spatiotemporal
Project Overview
- Tool to be developed: Optical microscopy-based tools to provide fast and facile predictions of the mode of entry of emerging pathogens.
- Tool to be developed: Optical microscopy protocols for simultaneous recording and correlation of standard and high resolution data of virus-membrane interactions.
- Tool to be developed: Machine learning-supported workflow in JIPipe for fast, objective, and quantitative image analysis and classification of virus-cell interactions.
Hypothesis enabled by the proposed tool: The prediction of the mode of virus uptake is important for choosing preventive measures of infections by emerging pathogens, and adaptation of optical microscopy and data analysis will provide the required tools.
Overarching CRC goals: Our project C03 couples high-spatiotemporal single-virus imaging with a machine-learning pipeline that infers entry modalities from standard widefield/confocal time-lapse data, enabling rapid, scalable annotation of membrane-interaction signatures across diverse viruses (G1, G3). Trained on growing consortium datasets, the classifier provides calibrated predictions of fusion vs. endocytic uptake routes for emerging pathogens, supporting early triage of infectivity-modifying interventions (G4).
Work Packages (WP):
- WP 1: Optimised visualisation of virus-membrane interactions (Eggeling/Schelhaas)
- WP 2: Fast prediction of virus entry modes through correlation of techniques (Eggeling/Figge/Schelhaas)
- WP 3: Machine-Learning-supported image analysis of virus-membrane interaction (Figge/Eggeling/SchelhaaS)
Team Members

Dr. Ziliang Zhao
Postdoc

Dr. Ruman Gerst
Postdoc

Ivan Avilov
Postdoc

Dr. Dhruv Khatri
Postdoc
2025
Brinkert, Pia; Krebs, Lena; Ventayol, Pilar Samperio; Greune, Lilo; Bannach, Carina; Amakiri, Cynthia; Bucher, Delia; Kollasser, Jana; Dersch, Petra; Boulant, Steeve; others,
The actin nucleation promoting factor WASH facilitates clathrin-independent endocytosis of human papillomaviruses Journal Article
In: EMBO reports, vol. 26, no. 22, pp. 5533, 2025.
@article{brinkert2025actin,
title = {The actin nucleation promoting factor WASH facilitates clathrin-independent endocytosis of human papillomaviruses},
author = {Pia Brinkert and Lena Krebs and Pilar Samperio Ventayol and Lilo Greune and Carina Bannach and Cynthia Amakiri and Delia Bucher and Jana Kollasser and Petra Dersch and Steeve Boulant and others},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {EMBO reports},
volume = {26},
number = {22},
pages = {5533},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2024
Dasgupta, Anindita; Koerfer, Agnes; Kokot, Boštjan; Urbančič, Iztok; Eggeling, Christian; Carravilla, Pablo
Effects and avoidance of photoconversion-induced artifacts in confocal and STED microscopy Journal Article
In: Nature Methods, vol. 21, no. 7, pp. 1171–1174, 2024.
@article{dasgupta2024effects,
title = {Effects and avoidance of photoconversion-induced artifacts in confocal and STED microscopy},
author = {Anindita Dasgupta and Agnes Koerfer and Boštjan Kokot and Iztok Urbančič and Christian Eggeling and Pablo Carravilla},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {Nature Methods},
volume = {21},
number = {7},
pages = {1171–1174},
publisher = {Nature Publishing Group US New York},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2023
Rizzato, Matteo; Mao, Fuxiang; Chardon, Florian; Lai, Kun-Yi; Villalonga-Planells, Ruth; Drexler, Hannes CA; Pesenti, Marion E; Fiskin, Mert; Roos, Nora; King, Kelly M; others,
Master mitotic kinases regulate viral genome delivery during papillomavirus cell entry Journal Article
In: Nature Communications, vol. 14, no. 1, pp. 355, 2023.
@article{rizzato2023master,
title = {Master mitotic kinases regulate viral genome delivery during papillomavirus cell entry},
author = {Matteo Rizzato and Fuxiang Mao and Florian Chardon and Kun-Yi Lai and Ruth Villalonga-Planells and Hannes CA Drexler and Marion E Pesenti and Mert Fiskin and Nora Roos and Kelly M King and others},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {Nature Communications},
volume = {14},
number = {1},
pages = {355},
publisher = {Nature Publishing Group UK London},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Svensson, Carl-Magnus; Reglinski, Katharina; Schliebs, Wolfgang; Erdmann, Ralf; Eggeling, Christian; Figge, Marc Thilo
Quantitative analysis of peroxisome tracks using a Hidden Markov Model Journal Article
In: Scientific reports, vol. 13, no. 1, pp. 19694, 2023.
@article{svensson2023quantitative,
title = {Quantitative analysis of peroxisome tracks using a Hidden Markov Model},
author = {Carl-Magnus Svensson and Katharina Reglinski and Wolfgang Schliebs and Ralf Erdmann and Christian Eggeling and Marc Thilo Figge},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {Scientific reports},
volume = {13},
number = {1},
pages = {19694},
publisher = {Nature Publishing Group UK London},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Gerst, Ruman; Cseresnyés, Zoltán; Figge, Marc Thilo
JIPipe: visual batch processing for ImageJ Journal Article
In: nature methods, vol. 20, no. 2, pp. 168–169, 2023.
@article{gerst2023jipipe,
title = {JIPipe: visual batch processing for ImageJ},
author = {Ruman Gerst and Zoltán Cseresnyés and Marc Thilo Figge},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {nature methods},
volume = {20},
number = {2},
pages = {168–169},
publisher = {Nature Publishing Group US New York},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2021
Lai, Kun-Yi; Rizzato, Matteo; Aydin, Inci; Villalonga-Planells, Ruth; Drexler, Hannes CA; Schelhaas, Mario
A Ran-binding protein facilitates nuclear import of human papillomavirus type 16 Journal Article
In: PLoS pathogens, vol. 17, no. 5, pp. e1009580, 2021.
@article{lai2021ran,
title = {A Ran-binding protein facilitates nuclear import of human papillomavirus type 16},
author = {Kun-Yi Lai and Matteo Rizzato and Inci Aydin and Ruth Villalonga-Planells and Hannes CA Drexler and Mario Schelhaas},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {PLoS pathogens},
volume = {17},
number = {5},
pages = {e1009580},
publisher = {Public Library of Science San Francisco, CA USA},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2019
Carravilla, Pablo; Chojnacki, Jakub; Rujas, Edurne; Insausti, Sara; Largo, Eneko; Waithe, Dominic; Apellaniz, Beatriz; Sicard, Taylor; Julien, Jean-Philippe; Eggeling, Christian; others,
Molecular recognition of the native HIV-1 MPER revealed by STED microscopy of single virions Journal Article
In: Nature communications, vol. 10, no. 1, pp. 78, 2019.
@article{carravilla2019molecular,
title = {Molecular recognition of the native HIV-1 MPER revealed by STED microscopy of single virions},
author = {Pablo Carravilla and Jakub Chojnacki and Edurne Rujas and Sara Insausti and Eneko Largo and Dominic Waithe and Beatriz Apellaniz and Taylor Sicard and Jean-Philippe Julien and Christian Eggeling and others},
year = {2019},
date = {2019-01-01},
urldate = {2019-01-01},
journal = {Nature communications},
volume = {10},
number = {1},
pages = {78},
publisher = {Nature Publishing Group UK London},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2018
Chojnacki, Jakub; Eggeling, Christian
Super-resolution fluorescence microscopy studies of human immunodeficiency virus Journal Article
In: Retrovirology, vol. 15, no. 1, pp. 41, 2018.
@article{chojnacki2018super,
title = {Super-resolution fluorescence microscopy studies of human immunodeficiency virus},
author = {Jakub Chojnacki and Christian Eggeling},
year = {2018},
date = {2018-01-01},
urldate = {2018-01-01},
journal = {Retrovirology},
volume = {15},
number = {1},
pages = {41},
publisher = {Springer},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2013
Mokhtari, Zeinab; Mech, Franziska; Zitzmann, Carolin; Hasenberg, Mike; Gunzer, Matthias; Figge, Marc Thilo
Automated characterization and parameter-free classification of cell tracks based on local migration behavior Journal Article
In: PloS one, vol. 8, no. 12, pp. e80808, 2013.
@article{mokhtari2013automated,
title = {Automated characterization and parameter-free classification of cell tracks based on local migration behavior},
author = {Zeinab Mokhtari and Franziska Mech and Carolin Zitzmann and Mike Hasenberg and Matthias Gunzer and Marc Thilo Figge},
year = {2013},
date = {2013-01-01},
urldate = {2013-01-01},
journal = {PloS one},
volume = {8},
number = {12},
pages = {e80808},
publisher = {Public Library of Science San Francisco, USA},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2012
Ewers, Helge; Schelhaas, Mario
Analysis of virus entry and cellular membrane dynamics by single particle tracking Journal Article
In: Methods in enzymology, vol. 506, pp. 63–80, 2012.
@article{ewers2012analysis,
title = {Analysis of virus entry and cellular membrane dynamics by single particle tracking},
author = {Helge Ewers and Mario Schelhaas},
year = {2012},
date = {2012-01-01},
urldate = {2012-01-01},
journal = {Methods in enzymology},
volume = {506},
pages = {63–80},
publisher = {Elsevier},
keywords = {},
pubstate = {published},
tppubtype = {article}
}


