
B03: Uncovering viral glycoprotein conformational dynamics for rational vaccine design

Projects of the CRC 1768

B03: Uncovering viral glycoprotein conformational dynamics for rational vaccine design
Structure-based prediction and design have made tremendous progress over the last thirty years with Rosetta and, especially, since 2021, when AlphaFold2 was released. This progress to achieve the holy grail of computational structural biology was rewarded with the Nobel Prize for these two algorithms in 2024. The accurate prediction of the protein structure is the first step towards an in silico first strategy to design vaccines and antibodies. Structure-based modelling will also provide estimates for host-receptor interactions, such as antibody-antigen complexes. With this information, the missing link between sequence and function can be filled. Here, we are developing methods to predict structures of viral glycoproteins that will allow us to assess emerging viral variants. With structure-based methods, we will investigate the impact of the observed mutations in viral glycoproteins on the structure and the respective conformational states. This is critical for understanding the conformational space that mediates the fusion process. One major step to overcome here is the lack of predictive power of AI tools for structure prediction to provide pre- and post-fusion receptor states. We will subsequently predict the effect these mutations have on the structure. Together with the consortium partners, we will investigate whether our methods predict the effect and the respective function of the viral glycoprotein (variants). The major virus-host interaction we will study is the interaction with the immune system, with the spike protein being the major target of the humoral response to SARS-CoV-2.
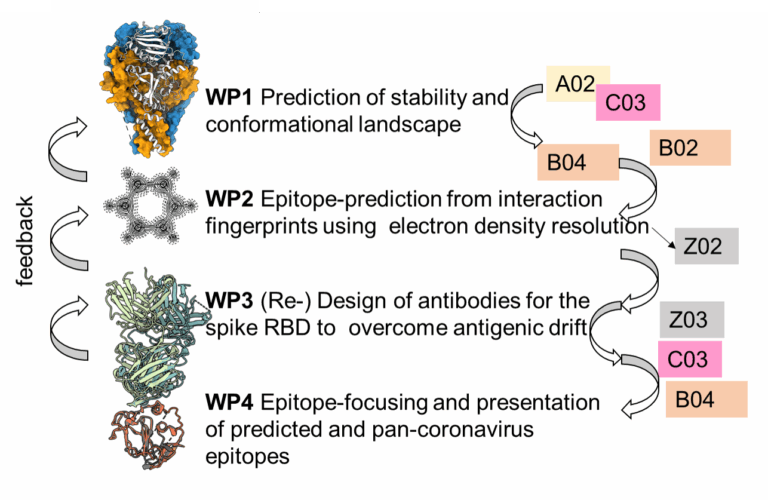
Antibodies, as major determinants of this immune response, are useful research tools and therapeutics but are challenging for structure prediction and design. Moreover, antigen-antibody interactions are inherently hard to predict and evaluate, even with emerging AI tools, due to the lack of data and the complexity of the molecular interaction. Here, we will overcome these limitations and develop a new tool termed ANNtibody that takes atomic and electron density calculations into account. Thus far, these methods could not be employed for systems with more than a couple dozen atoms, but the training of AI on electron density data or on Density Functional Theory (DFT) calculations circumvents these resource limited steps. Data from NFDI4Chem and its associated repositories will be used to benchmark these.
With this increase in resolution, we hypothesise that our method will capture the complex interaction network in the antibody-antigen interface more accurately. These calculations will be used to predict antibody- antigen interactions, which we will challenge with the experimental design of antibodies for emerging SARS- CoV-2 variants using antibody interactions with the highly variable receptor-binding domain. With our method, we will update these antibody sequences and test experimentally whether we can overcome viral es- cape. Subsequently, we will design epitope focused immunogens based on SARS-CoV-2 epitopes that will elicit broadly neutralising antibody populations. All experimentally obtained data will be used to refine the developed methods. Data provided by C02 (Deckert/Deinhardt/Emmer) on virus-host interactions, by A02 (Friedel/Kühnert) and NFDI4Microbiota on viral sequences, and by B04 (Dittrich/McHardy) on surveillance will be essential for training and optimising our tool. These datasets will be integrated with our structural features, enabling our tool to generate predictions that will directly inform and support our partner projects. Altogether, we will generate computational structure-based tools that will help answers goals G1 and G3, providing insight into the host-receptor interaction. These tools will allow us to rapidly fight emerging viral infections and tune these tools for vaccine design, probing our ability to generate a broadly neutralising vaccine.
Project Overview
Design of structure-based tools that accurately predict viral glycoproteins and subsequently their interactions with antibodies, with atomic resolution. Integration of the electron density data will overcome current limitations in the accuracy of interaction predictions. These technologies will be evaluated using the design of antibodies and epitope-focused immunogens.
Our structural models will be able to link sequence data from A02 (Friedel/Kühnert) and phenotype data from B04 (Dittrich/McHardy) and subsequently more CRC partners through modelling of the antigen structures and their antigen-antibody interactions. This will be a major cornerstone of the CRC’s aim to integrate bioinformatic tools for virology.
With the computational methods developed here and tested together with B04 (Dittrich/McHardy), we are contributing to the CRC VirusREvolution overarching question on tools for novel emerging viruses and their variants. With our structure-based modelling of the conformational landscape of the virus envelope protein and the prediction of antibody-antigen interactions, we will be able to predict the effect of observed mutations in novel emerging viral variants on antibody and vaccine effectiveness. These tools further allow us to profile virus-host interactions with a special focus on virus immune system interactions, allowing us to predict escape and update currently existing antibody therapeutics to provide protection against new emerging variants. Subsequently, we will provide proof- of-principle that computational methods are able to inform vaccine design that will not be sensitive to escape because of targeting conserved, broadly neutralising epitopes using epitope information with atomic resolution. These vaccines will be produced and experimentally tested together with partners in the CRC VirusREvolution to establish a computational-experimental feedback loop. In case of an outbreak of an emerging viral disease, our tools will be used to design vaccines that elicit the most important antibody responses for protection.
- Tool to be developed: Optimised tools for glycoprotein conformational state prediction and the prediction of antibody- antigen interactions with atom resolution to inform epitope-focused immunogen and antibody design.
Hypothesis enabled by the proposed tool: High-resolution prediction of antibody-antigen interaction will overcome current limitations in antibody and antigen design to inform vaccine development.
Overarching CRC goals: Our project B03 develops structure-based pipelines to predict and interrogate conformational dynamics of viral glycoproteins under mutation and introduces ANNtibody, an electron-density-informed AI for modelling antibody-antigen interfaces and designing epitope-focused immunogens. This tool enables rapid assessment of emerging variants and immune interactions in surveillance context (G1, G2). Benchmarking across SARS-CoV-2, RVFV and related group IV/V viruses yields generalisable principles linking glycoprotein conformational landscapes to fusion and neutralization, providing transferable rules of virus-host (immune) interaction (G3).
Work Packages (WP):
- WP 1: Prediction of stability and conformational landscape of viral glycoprotein with artificial-intelligence tools taking antigenic drift into consideration (Meiler, Schoeder)
- WP 2: Epitope prediction from interaction fingerprints using electron density resolution (Meiler)
- WP 3: (Re-) Design of antibodies for the SARS-CoV-2 spike RBD to overcome antigenic drift (Schoeder, Meiler)
- WP 4: Epitope-focusing and presentation of predicted and pan-coronavirus epitopes (Schoeder)
Team Members

PhD B03 1
PhD Student

PhD B03 2
PhD Student
2025
Ertelt, Moritz; Moretti, Rocco; Meiler, Jens; Schoeder, Clara T
Self-supervised machine learning methods for protein design improve sampling but not the identification of high-fitness variants. Journal Article
In: Science advances, vol. 11, iss. 7, pp. eadr7338, 2025.
@article{Ertelt:25,
title = {Self-supervised machine learning methods for protein design improve sampling but not the identification of high-fitness variants.},
author = {Moritz Ertelt and Rocco Moretti and Jens Meiler and Clara T Schoeder},
url = {https://pubmed.ncbi.nlm.nih.gov/39937901/},
doi = {10.1126/sciadv.adr7338},
year = {2025},
date = {2025-02-01},
journal = {Science advances},
volume = {11},
issue = {7},
pages = {eadr7338},
keywords = {},
pubstate = {ppublish},
tppubtype = {article}
}
2024
Ertelt, Moritz; Meiler, Jens; Schoeder, Clara T
Combining Rosetta sequence design with protein language model predictions using evolutionary scale modeling (ESM) as restraint. Journal Article
In: ACS synthetic biology, vol. 13, iss. 4, no. 4, pp. 1085–1092, 2024.
@article{Ertelt:24:rosetta,
title = {Combining Rosetta sequence design with protein language model predictions using evolutionary scale modeling (ESM) as restraint.},
author = {Moritz Ertelt and Jens Meiler and Clara T Schoeder},
url = {https://pubmed.ncbi.nlm.nih.gov/38568188/},
doi = {10.1021/acssynbio.3c00753},
year = {2024},
date = {2024-04-01},
journal = {ACS synthetic biology},
volume = {13},
number = {4},
issue = {4},
pages = {1085–1092},
keywords = {},
pubstate = {ppublish},
tppubtype = {article}
}
Ertelt, Moritz; Mulligan, Vikram Khipple; Maguire, Jack B; Lyskov, Sergey; Moretti, Rocco; Schiffner, Torben; Meiler, Jens; Schoeder, Clara T
Combining machine learning with structure-based protein design to predict and engineer post-translational modifications of proteins. Journal Article
In: PLoS Comput Biol, vol. 20, iss. 3, no. 3, pp. e1011939, 2024.
@article{Ertelt:24:machine,
title = {Combining machine learning with structure-based protein design to predict and engineer post-translational modifications of proteins.},
author = {Moritz Ertelt and Vikram Khipple Mulligan and Jack B Maguire and Sergey Lyskov and Rocco Moretti and Torben Schiffner and Jens Meiler and Clara T Schoeder},
url = {https://pubmed.ncbi.nlm.nih.gov/38484014/},
doi = {10.1371/journal.pcbi.1011939},
year = {2024},
date = {2024-03-01},
journal = {PLoS Comput Biol},
volume = {20},
number = {3},
issue = {3},
pages = {e1011939},
publisher = {Public Library of Science (PLoS)},
keywords = {},
pubstate = {epublish},
tppubtype = {article}
}
2023
Sala, D; Engelberger, F; Mchaourab, H S; Meiler, J
Modeling conformational states of proteins with AlphaFold Journal Article
In: Curr Opin Struct Biol, vol. 81, pp. 102645, 2023.
@article{Sala:23,
title = {Modeling conformational states of proteins with AlphaFold},
author = {D Sala and F Engelberger and H S Mchaourab and J Meiler},
url = {https://www.sciencedirect.com/science/article/pii/S0959440X23001197},
doi = {10.1016/j.sbi.2023.102645},
year = {2023},
date = {2023-08-01},
journal = {Curr Opin Struct Biol},
volume = {81},
pages = {102645},
keywords = {},
pubstate = {ppublish},
tppubtype = {article}
}
Sala, Davide; Hildebrand, Peter W; Meiler, Jens
Biasing AlphaFold2 to predict GPCRs and kinases with user-defined functional or structural properties Journal Article
In: Front Mol Biosci, vol. 10, pp. 1121962, 2023.
@article{Sala:23:biasing,
title = {Biasing AlphaFold2 to predict GPCRs and kinases with user-defined functional or structural properties},
author = {Davide Sala and Peter W Hildebrand and Jens Meiler},
url = {https://www.frontiersin.org/articles/10.3389/fmolb.2023.1121962},
doi = {10.3389/fmolb.2023.1121962},
year = {2023},
date = {2023-01-01},
journal = {Front Mol Biosci},
volume = {10},
pages = {1121962},
keywords = {},
pubstate = {epublish},
tppubtype = {article}
}
2022
Schoeder, Clara T; Gilchuk, Pavlo; Sangha, Amandeep K; Ledwitch, Kaitlyn V; Malherbe, Delphine C; Zhang, Xuan; Binshtein, Elad; Williamson, Lauren E; Martina, Cristina E; Dong, Jinhui; Armstrong, Erica; Sutton, Rachel; Nargi, Rachel; Rodriguez, Jessica; Kuzmina, Natalia; Fiala, Brooke; King, Neil P; Bukreyev, Alexander; Crowe, James E; Meiler, Jens
Epitope-focused immunogen design based on the ebolavirus glycoprotein HR2-MPER region Journal Article
In: PLoS Pathog, vol. 18, iss. 5, no. 5, pp. 1-29, 2022.
@article{Schoeder:22,
title = {Epitope-focused immunogen design based on the ebolavirus glycoprotein HR2-MPER region},
author = {Clara T Schoeder and Pavlo Gilchuk and Amandeep K Sangha and Kaitlyn V Ledwitch and Delphine C Malherbe and Xuan Zhang and Elad Binshtein and Lauren E Williamson and Cristina E Martina and Jinhui Dong and Erica Armstrong and Rachel Sutton and Rachel Nargi and Jessica Rodriguez and Natalia Kuzmina and Brooke Fiala and Neil P King and Alexander Bukreyev and James E Crowe and Jens Meiler},
editor = {Thomas Hoenen},
url = {https://doi.org/10.1371/journal.ppat.1010518},
doi = {10.1371/journal.ppat.1010518},
year = {2022},
date = {2022-05-01},
journal = {PLoS Pathog},
volume = {18},
number = {5},
issue = {5},
pages = {1-29},
publisher = {Public Library of Science},
keywords = {},
pubstate = {epublish},
tppubtype = {article}
}
Alamo, Diego Del; Sala, Davide; Mchaourab, Hassane S; Meiler, Jens
Sampling alternative conformational states of transporters and receptors with AlphaFold2 Journal Article
In: eLife, vol. 11, pp. e75751, 2022.
@article{Alamo:22,
title = {Sampling alternative conformational states of transporters and receptors with AlphaFold2},
author = {Diego Del Alamo and Davide Sala and Hassane S Mchaourab and Jens Meiler},
editor = {Janice L Robertson and Kenton J Swartz and Janice L Robertson},
url = {https://doi.org/10.7554/eLife.75751},
doi = {10.7554/eLife.75751},
year = {2022},
date = {2022-03-01},
journal = {eLife},
volume = {11},
pages = {e75751},
publisher = {eLife Sciences Publications, Ltd},
keywords = {},
pubstate = {epublish},
tppubtype = {article}
}
2021
Schoeder, Clara T; Schmitz, Samuel; Adolf-Bryfogle, Jared; Sevy, Alexander M; Finn, Jessica A; Sauer, Marion F; Bozhanova, Nina G; Mueller, Benjamin K; Sangha, Amandeep K; Bonet, Jaume; Sheehan, Jonathan H; Kuenze, Georg; Marlow, Brennica; Smith, Shannon T; Woods, Hope; Bender, Brian J; Martina, Cristina E; Alamo, Diego Del; Kodali, Pranav; Gulsevin, Alican; Schief, William R; Correia, Bruno E; Crowe, James E; Meiler, Jens; Moretti, Rocco
Modeling immunity with Rosetta: Methods for antibody and antigen design Journal Article
In: Biochemistry, vol. 60, iss. 11, pp. 825–846, 2021.
@article{Schoeder:21,
title = {Modeling immunity with Rosetta: Methods for antibody and antigen design},
author = {Clara T Schoeder and Samuel Schmitz and Jared Adolf-Bryfogle and Alexander M Sevy and Jessica A Finn and Marion F Sauer and Nina G Bozhanova and Benjamin K Mueller and Amandeep K Sangha and Jaume Bonet and Jonathan H Sheehan and Georg Kuenze and Brennica Marlow and Shannon T Smith and Hope Woods and Brian J Bender and Cristina E Martina and Diego Del Alamo and Pranav Kodali and Alican Gulsevin and William R Schief and Bruno E Correia and James E Crowe and Jens Meiler and Rocco Moretti},
doi = {10.1021/acs.biochem.0c00912},
year = {2021},
date = {2021-01-01},
journal = {Biochemistry},
volume = {60},
issue = {11},
pages = {825–846},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2020
Alexander, Matthew R; Schoeder, Clara T; Brown, Jacquelyn A; Smart, Charles D; Moth, Chris; Wikswo, John P; Capra, John A; Meiler, Jens; Chen, Wenbiao; Madhur, Meena S
Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species Journal Article
In: The FASEB Journal, vol. 34, iss. 12, no. 12, pp. 15946-15960, 2020.
@article{Alexander:20,
title = {Predicting susceptibility to SARS-CoV-2 infection based on structural differences in ACE2 across species},
author = {Matthew R Alexander and Clara T Schoeder and Jacquelyn A Brown and Charles D Smart and Chris Moth and John P Wikswo and John A Capra and Jens Meiler and Wenbiao Chen and Meena S Madhur},
url = {https://pubmed.ncbi.nlm.nih.gov/33015868/},
doi = {10.1096/fj.202001808R},
year = {2020},
date = {2020-12-01},
journal = {The FASEB Journal},
volume = {34},
number = {12},
issue = {12},
pages = {15946-15960},
publisher = {Wiley},
keywords = {},
pubstate = {ppublish},
tppubtype = {article}
}
Sauer, Marion F; Sevy, Alexander M; Crowe, James E; Meiler, Jens
Multi-state design of flexible proteins predicts sequences optimal for conformational change Journal Article
In: PLoS Comput Biol, vol. 16, iss. 2, pp. e1007339, 2020.
@article{Sauer:20,
title = {Multi-state design of flexible proteins predicts sequences optimal for conformational change},
author = {Marion F Sauer and Alexander M Sevy and James E Crowe and Jens Meiler},
url = {https://pubmed.ncbi.nlm.nih.gov/32032348/},
doi = {10.1371/journal.pcbi.1007339},
year = {2020},
date = {2020-02-01},
journal = {PLoS Comput Biol},
volume = {16},
issue = {2},
pages = {e1007339},
publisher = {Public Library of Science (PLoS)},
keywords = {},
pubstate = {epublish},
tppubtype = {article}
}


