
Z02: INFormation, INFormatics, and INFrastructure

Projects of the CRC 1768

Z02: INFormation, INFormatics, and INFrastructure
Central activities of the CRC VirusREvolution
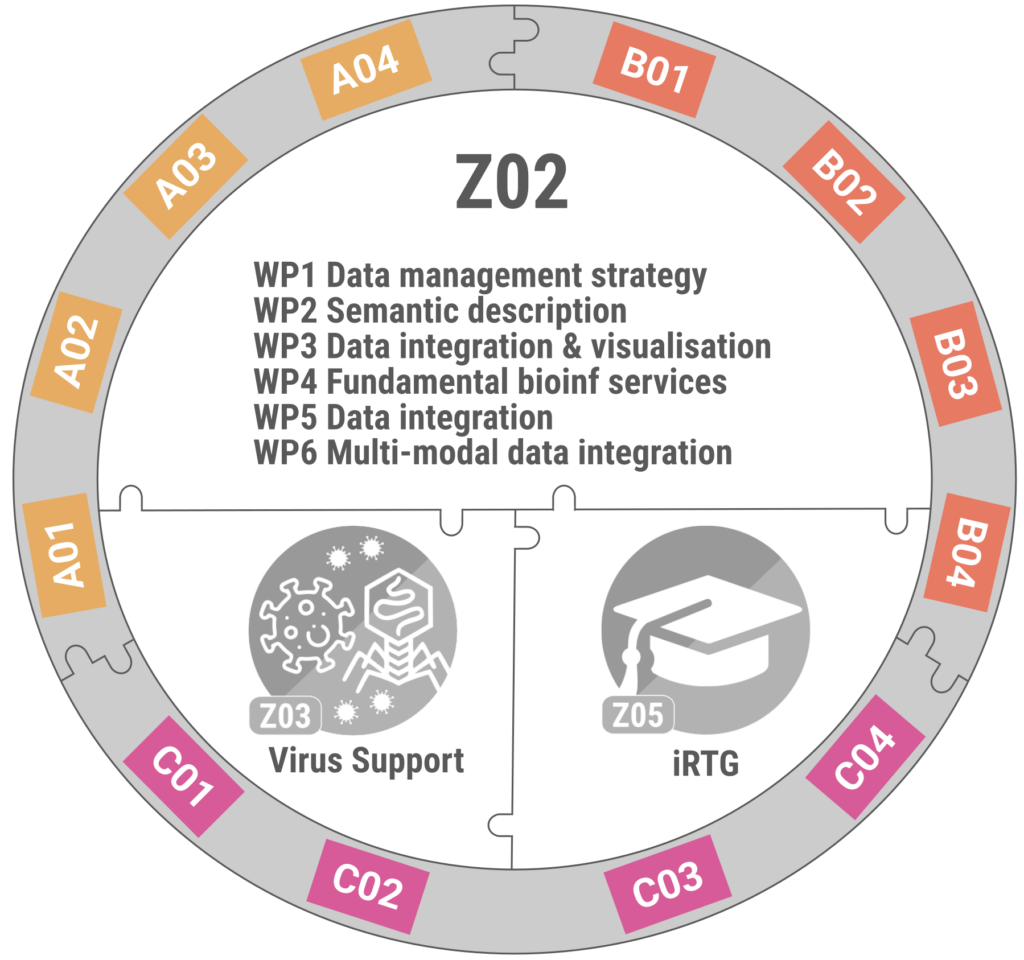
Overview of the Z02 INF project plan.
For example, as outlined below, NFDI-supported tools and repositories will be used as appropriate. In the following, we describe the state of the art and our own preliminary work in three key fields for this project: data management, bioinformatics services, and data integration.
In large, data-intensive collaborative projects, it has been shown that centrally organised data management contributes significantly to the success of a project. The data management team often implements and operates the necessary technical infrastructure for data storage and exchange. It curates data to ensure quality and fulfil the requirements of the above-mentioned stakeholders. It supports researchers in using the infrastructure together with the project participants. It develops policies and recommendations for uniform and sustainable data management in the project. Examples of this are the data management in the DFG priority programme Biodiversity Exploratories or the Jena Cluster of Excellence Balance of the Microverse.
e. Data integration in the VirJenDB – state of the art. Metadata, or data that describes primary data, plays a central role in integrating results across different omics techniques. This integration can be achieved through the application of controlled vocabularies from ontologies, the use of common databases for annotation, and leveraging established file formats for different data types. For sequence data deposition, repositories such as the European Nucleotide Archive (ENA) and the NCBI GenBank adopt open, community-driven standards to model submitted datasets. For example, the most relevant community-driven standards for virus research include the Genome Standards Consortium (GSC), Minimum Information about an Uncultivated Virus Genome (MIUViG), and Minimal Information about a Genome Sequence: Virus (MIGSVi) packages. Moreover, minimal information standards have been established for transcriptomics, proteomics, and metabolomics data, such as the MIAME (Minimum Information About a Microarray Experiment) and mzML. Shared ontology resources providing additional semantic structure across virus omics datasets include the Gene Ontology (GO), IntEnz, KEGG-Virus, PDBeChem, ChEMBL, Disease Ontology, and Virus Orthologous Groups (VOGs). These standards organise metadata into modular categories such as sample collection, protocols, consent, and provenance, which can then be linked via permanent identifiers. Such linkage enables the integration of genomics, transcriptomics, proteomics, and metabolomics datasets in cross-disciplinary projects. Integration across project, experiment, and sample levels can be demonstrated by data management tools such as the NFDI-supported biodiversity platform BEXIS2.
The VirJenDB provides a user-friendly web portal developed with continuous feedback from the virus research community via surveys at conferences and personal interviews. Major features of the portal include search, browse, and API access to the dataset of sequences, clusters, and metadata. ElasticSearch provides a powerful search engine while RestAPI and SwaggerUI allow for programmatic access. Further, summary statistics and visualisations are continuously improved. For the CRC, a private workbench is being developed to provide access to a project-specific management and analysis environment, including support for data integration and exploratory visualisations of the CRC tool outputs. The VirJenDB platform is intended to focus community curation efforts and enrich repository submissions of cross-disciplinary virus datasets through extending genomic standards, thereby bridging the eukaryotic and bacteriophage research communities. These aligned aims and features make the NFDI4Microbiota database the ideal infrastructure hub to integrate the cross-disciplinary CRC research outputs and to streamline the unification of the CRC tools in the third funding period.
Project Overview
- WP 1 and WP 2 establish the CRC-wide data management strategy and semantic metadata frameworks.
- WP 3 extends the VirJenDB portal and develops it towards a central integration and access point for virus data generated in the CRC.
- WP 4 provides core bioinformatic services and transforms prototype tools into usable pipelines.
- WP 5 delivers curated reference genomes and annotations for all viruses used in the CRC VirusREvolution.
- WP 6 integrates multi-omics and spectral data to generate predictive models of virus-host interaction.
Work Packages (WP):
- WP 1: Data management strategy (Gerlach/König-Ries)
- WP 2: Semantic description and publication of research objects (König-Ries/Gerlach/Cassman)
- WP 3: Connecting CRC-specific data, tools, and visualisations within VirJenDB (Cassman)
- WP 4: Fundamental bioinformatic services (Barth)
- WP 5: Bioinformatics workflows for the model viruses (Barth/Cassman)
- WP 6: Integration of transcriptomics, metabolomics, and proteomics data of the model viruses (Barth/Cassman)
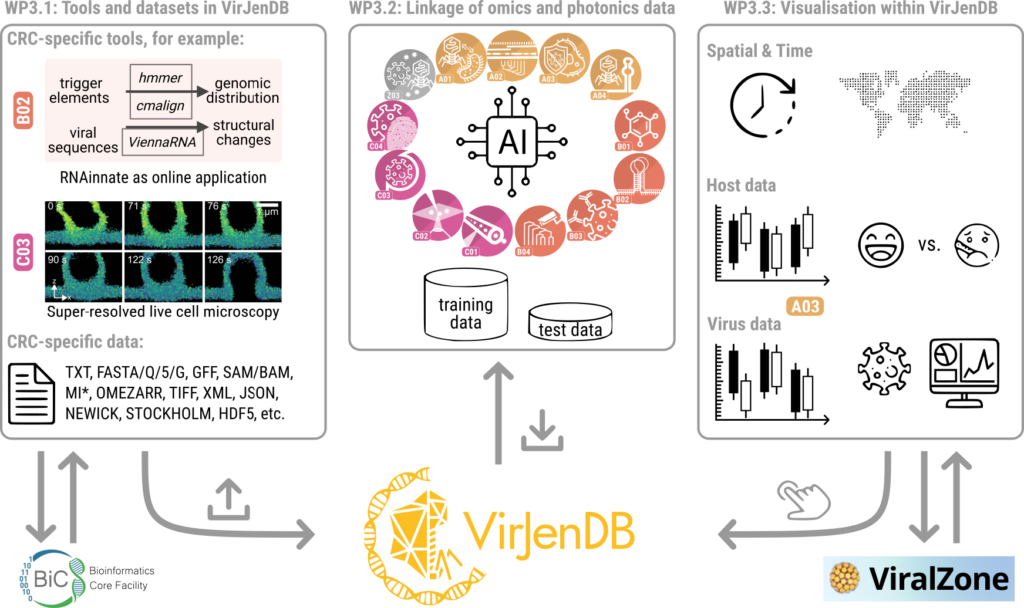
WP 3 workflow for data integration and implementation in the VirJenDB.
Team Members

PhD Z01 1
PhD Student
2025
Saghaei, Shahram; Siemers, Malte; Ossetek, Kilian L; Richter, Stephan; Edwards, Robert A; Roux, Simon; Zielezinski, Andrzej; Dutilh, Bas E; Marz, Manja; Cassman, Noriko A
VirJenDB: a FAIR (meta) data and bioinformatics platform for all viruses Journal Article
In: Nucleic Acids Research, pp. gkaf1224, 2025.
@article{saghaei2025virjendb,
title = {VirJenDB: a FAIR (meta) data and bioinformatics platform for all viruses},
author = {Shahram Saghaei and Malte Siemers and Kilian L Ossetek and Stephan Richter and Robert A Edwards and Simon Roux and Andrzej Zielezinski and Bas E Dutilh and Manja Marz and Noriko A Cassman},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Nucleic Acids Research},
pages = {gkaf1224},
publisher = {Oxford University Press},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2023
Ritsch, Muriel; Cassman, Noriko A; Saghaei, Shahram; Marz, Manja
Navigating the landscape: a comprehensive review of current virus databases Journal Article
In: Viruses, vol. 15, no. 9, pp. 1834, 2023.
@article{ritsch2023navigating,
title = {Navigating the landscape: a comprehensive review of current virus databases},
author = {Muriel Ritsch and Noriko A Cassman and Shahram Saghaei and Manja Marz},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {Viruses},
volume = {15},
number = {9},
pages = {1834},
publisher = {MDPI},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2021
Mock, Florian; Viehweger, Adrian; Barth, Emanuel; Marz, Manja
VIDHOP, viral host prediction with deep learning Journal Article
In: Bioinformatics, vol. 37, no. 3, pp. 318–325, 2021.
@article{mock2021vidhop,
title = {VIDHOP, viral host prediction with deep learning},
author = {Florian Mock and Adrian Viehweger and Emanuel Barth and Manja Marz},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Bioinformatics},
volume = {37},
number = {3},
pages = {318–325},
publisher = {Oxford University Press},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Chamanara, Javad; Gaikwad, Jitendra; Gerlach, Roman; Algergawy, Alsayed; Ostrowski, Andreas; König-Ries, Birgitta
BEXIS2: A FAIR-aligned data management system for biodiversity, ecology and environmental data Journal Article
In: Biodiversity Data Journal, vol. 9, pp. e72901, 2021.
@article{chamanara2021bexis2,
title = {BEXIS2: A FAIR-aligned data management system for biodiversity, ecology and environmental data},
author = {Javad Chamanara and Jitendra Gaikwad and Roman Gerlach and Alsayed Algergawy and Andreas Ostrowski and Birgitta König-Ries},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Biodiversity Data Journal},
volume = {9},
pages = {e72901},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Samuel, Sheeba; König-Ries, Birgitta
Understanding experiments and research practices for reproducibility: an exploratory study Journal Article
In: PeerJ, vol. 9, pp. e11140, 2021.
@article{samuel2021understanding,
title = {Understanding experiments and research practices for reproducibility: an exploratory study},
author = {Sheeba Samuel and Birgitta König-Ries},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {PeerJ},
volume = {9},
pages = {e11140},
publisher = {PeerJ Inc.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Löffler, Felicitas; Wesp, Valentin; König-Ries, Birgitta; Klan, Friederike
Dataset search in biodiversity research: Do metadata in data repositories reflect scholarly information needs? Journal Article
In: PloS one, vol. 16, no. 3, pp. e0246099, 2021.
@article{loffler2021dataset,
title = {Dataset search in biodiversity research: Do metadata in data repositories reflect scholarly information needs?},
author = {Felicitas Löffler and Valentin Wesp and Birgitta König-Ries and Friederike Klan},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {PloS one},
volume = {16},
number = {3},
pages = {e0246099},
publisher = {Public Library of Science San Francisco, CA USA},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2020
Srivastava, Akash; Barth, Emanuel; Ermolaeva, Maria A; Guenther, Madlen; Frahm, Christiane; Marz, Manja; Witte, Otto W
Tissue-specific gene expression changes are associated with aging in mice Journal Article
In: Genomics, proteomics & bioinformatics, vol. 18, no. 4, pp. 430–442, 2020.
@article{srivastava2020tissue,
title = {Tissue-specific gene expression changes are associated with aging in mice},
author = {Akash Srivastava and Emanuel Barth and Maria A Ermolaeva and Madlen Guenther and Christiane Frahm and Manja Marz and Otto W Witte},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {Genomics, proteomics & bioinformatics},
volume = {18},
number = {4},
pages = {430–442},
publisher = {Oxford University Press},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2019
Barth, Emanuel; Srivastava, Akash; Stojiljkovic, Milan; Frahm, Christiane; Axer, Hubertus; Witte, Otto W; Marz, Manja
Conserved aging-related signatures of senescence and inflammation in different tissues and species Journal Article
In: Aging (Albany NY), vol. 11, no. 19, pp. 8556, 2019.
@article{barth2019conserved,
title = {Conserved aging-related signatures of senescence and inflammation in different tissues and species},
author = {Emanuel Barth and Akash Srivastava and Milan Stojiljkovic and Christiane Frahm and Hubertus Axer and Otto W Witte and Manja Marz},
year = {2019},
date = {2019-01-01},
urldate = {2019-01-01},
journal = {Aging (Albany NY)},
volume = {11},
number = {19},
pages = {8556},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2012
Cassman, Noriko; Prieto-Davó, Alejandra; Walsh, Kevin; Silva, Genivaldo GZ; Angly, Florent; Akhter, Sajia; Barott, Katie; Busch, Julia; McDole, Tracey; Haggerty, J Matthew; others,
Oxygen minimum zones harbour novel viral communities with low diversity Journal Article
In: Environmental microbiology, vol. 14, no. 11, pp. 3043–3065, 2012.
@article{cassman2012oxygen,
title = {Oxygen minimum zones harbour novel viral communities with low diversity},
author = {Noriko Cassman and Alejandra Prieto-Davó and Kevin Walsh and Genivaldo GZ Silva and Florent Angly and Sajia Akhter and Katie Barott and Julia Busch and Tracey McDole and J Matthew Haggerty and others},
year = {2012},
date = {2012-01-01},
urldate = {2012-01-01},
journal = {Environmental microbiology},
volume = {14},
number = {11},
pages = {3043–3065},
publisher = {Wiley Online Library},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
0000
Axtmann, Alexandra
”Wir bringen die breite Basis mit” – Gemeinsames Plädoyer für eine enge Einbindung der Landesinitiativen für Forschungsdatenmanagement in die Nationale Forschungsdateninfrastruktur. Journal Article
In: 0000.
@article{axtmannwir,
title = {”Wir bringen die breite Basis mit” – Gemeinsames Plädoyer für eine enge Einbindung der Landesinitiativen für Forschungsdatenmanagement in die Nationale Forschungsdateninfrastruktur.},
author = {Alexandra Axtmann},
publisher = {Deutsche Nationalbibliothek},
keywords = {},
pubstate = {published},
tppubtype = {article}
}


