
B02: RNA determinants of the antiviral innate immune response

Projects of the CRC 1768

B02: RNA determinants of the antiviral innate immune response
Viral RNAs trigger the innate immune response, which is able not only to distinguish foreign RNA from the cell’s own diverse complement of RNA molecules, but also varies among viral pathogens. Macrophages act as the first line of defence against invading viruses and are pivotal for the innate immune response, crucially regulating the entire inflammatory process during viral infections. Single- or double-stranded RNA viruses are recognised by several classes of RNA-binding proteins, in particular, the RNA-dependent toll-like receptors (TLR), TLR-7 and TLR-8, the RIG-I-like receptors (RLR) RIG-I, MDA5, LGP2, and their homologues, and the double-strand sensing protein kinase R (PKR). While individual RNA ligands of these sensory proteins have been well studied, a comprehensive and comparative understanding of their evolutionary and structural diversity across viral groups remains lacking.
In our project, we therefore aim to determine systematically the key features of both RNA sequence and RNA structure required for binding to these sensory proteins and for the subsequent activation of the innate immune response. We aim to make this connection actionable by developing a comprehensive predictive toolkit, RNAinnate, designed to predict the tempo and mode of activation of the innate immune system from the viral RNA sequence.
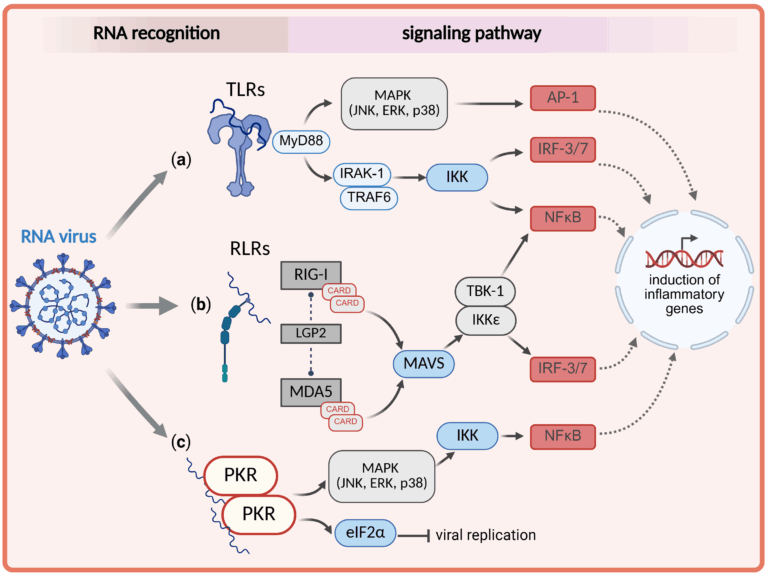
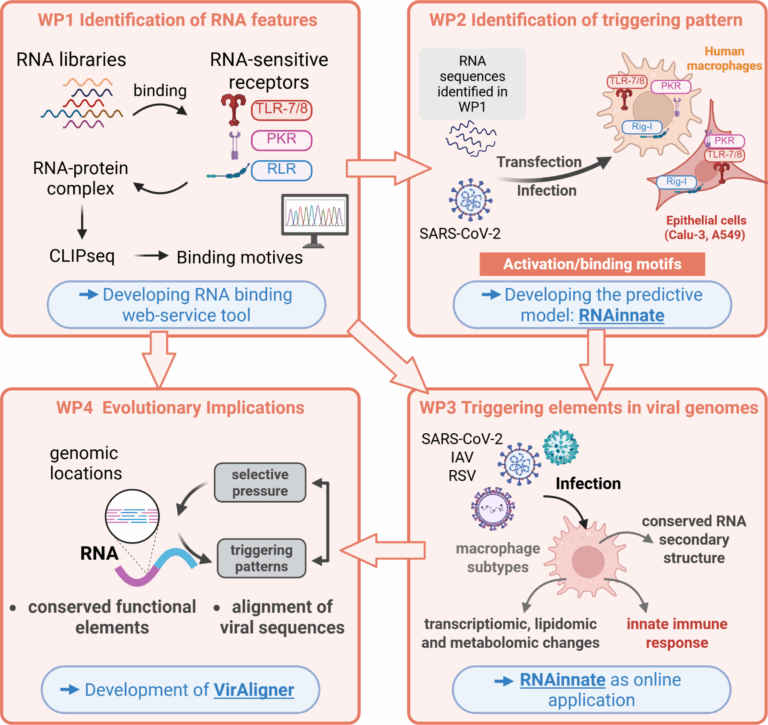
In detail, we will first investigate the specificity of RNA-protein binding for each purified RNA sensor in vitro, using randomised synthesised RNA libraries and employing cross-linking and immunoprecipitation sequencing methods (CLIP-seq). We will then transfect cells with distinct RNA sequences to validate our findings on the purified RNA sensors using electroporation- and DharmaFECT-based transfection. We will establish a transfection protocol using human lung epithelial cells (A549 cells and Calu-3 cells) and subsequently apply this technique to human primary macrophages. We will thoroughly examine the innate immune response, in particular the subsequent phosphorylation of downstream kinases of the specific RNA-binding proteins in the lung epithelial cells and in the macrophages. Afterwards, we will infect lung epithelial cells and human macrophages with SARS-CoV-2 to investigate virus-specific RNA-protein binding. Ultimately, we will infect cells in parallel with intact SARS-CoV-2, influenza A virus (IAV), and respiratory syncytial virus (RSV) to map the previously identified triggering elements within their natural genomic contexts. In order to determine characteristics of binding RNAs, we will evaluate enrichment and depletion of features such as secondary structure elements and local sequence motifs, as well as assess the distribution of folding energies. Moreover, we will employ unsupervised clustering techniques on these features, and combine the results of the different methods to extract descriptors of binding patterns in the form of covariance models and Bayesian descriptors similar to Dimont. Besides the determination of the RNA sequences, we will analyse the transcriptomic, metabolomic, and lipidomic changes of human primary macrophages related to the specific RNA sensors that occur following distinct RNA recognition. Finally, we will uncover evidence of selection pressure that removes or attenuates the individual RNA trigger elements for the innate immune system, providing insights into the mechanisms of RNA genome evolution. To achieve the latter goal, scalable, high-quality alignments of viral genomes are required that can combine inter-species comparison with information of strain-level variations. As no such tool exists, we fill this gap with VirAligner, using novel combinations of existing approaches in comparative sequence analysis.
Project Overview
We aim to understand the sequence and structure requirements of viral RNAs that are necessary to trigger the innate immune system, and to disentangle the reasons for differences in the response. This overarching biological question requires that we tackle the following research tasks:
- What are the sequence and structure motifs of the RNAs that bind to the different sensory proteins (RNA-dependent toll-like receptors, RIG-I-like receptors, and PKR)?
- How does the interplay of different binders contribute to the activation of key pathways in the innate immunesystem?
- Can the activation patterns be predicted from the viral RNA sequence and/or the knowledge of viral transcripts?
- Does the interaction of the viral RNAs with the sensory proteins of the innate immune system lead to detectable evolutionary constraints, such as the systematic avoidance of high-affinity binding patterns?
In order to successfully address these questions, several technical challenges have to be overcome:
- We aim to construct models that predict binding propensities for the sensory proteins as a function of the binding RNA.
- The (predicted) binding patterns will need to be aggregated to predict the pattern of the immune response.
- The analysis of the evolutionary effects requires high-quality alignments of entire viral genomes
- Tool to be developed:
VirAligner, a high-quality aligner for viral sequences, using graph theoretic, anchor-based, and dynamic-programming methods to handle sequence conservation variations, genomic rearrangements, and incomplete sequences. - Tool to be developed:
RNAinnate, a learning-based prediction tool for the stimulation of the innate immune system by viral RNAs.
Hypothesis enabled by the proposed tool: The specificity of the innate immune response to viral RNAs derives from
the interplay of multiple pathways rather than a single trigger.
Overarching CRC goals: Our project develops VirAligner for scalable, rearrangement-tolerant whole-genome alignments and RNAinnate, a learning-based toolkit that links viral RNA sequence/structure features to binding and activation of innate RNA sensors (RIG-I/MDA5/LGP2, TLR7/8, PKR), enabling precise description of innate trigger elements across viruses (G1). By integrating CLIP-seq, designed RNA libraries, and infection assays in epithelial cells and primary macrophages, the project derives generalisable rules of RNA-mediated sensing and reveals evolutionary selection acting on trigger motifs across species and strains (G2, G3).
Work Packages (WP):
- WP 1: Identification RNA features (Jordan/Stadler)
- WP 2: Identification of triggering patterns to activate innate immune response (Jordan/Stadler)
- WP 3: Triggering elements in viral genomes (Jordan/Stadler)
- WP 4: Evolutionary Implications (Stadler)
- WP 5: Identifying antiviral treatment strategies in the event of pandemic outbreak (Jordan)
Team Members
2025
Hornung, Franziska; SureshKumar, Harini K; Klement, Laura; Reisser, Yasmina; Wernike, Christoph; Nischang, Vivien; Jordan, Paul M; Werz, Oliver; Hoffmann, Carsten; Löffler, Bettina; others,
High-fat diet impairs microbial metabolite production and aggravates influenza A infection Journal Article
In: Cell Communication and Signaling, vol. 23, no. 1, pp. 359, 2025.
@article{hornung2025high,
title = {High-fat diet impairs microbial metabolite production and aggravates influenza A infection},
author = {Franziska Hornung and Harini K SureshKumar and Laura Klement and Yasmina Reisser and Christoph Wernike and Vivien Nischang and Paul M Jordan and Oliver Werz and Carsten Hoffmann and Bettina Löffler and others},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Cell Communication and Signaling},
volume = {23},
number = {1},
pages = {359},
publisher = {Springer},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2024
Löhneysen, Sarah; Spicher, Thomas; Varenyk, Yuliia; Yao, Hua-Ting; Lorenz, Ronny; Hofacker, Ivo; Stadler, Peter F
Phylogenetic and chemical probing information as soft constraints in RNA secondary structure prediction. Journal Article
In: Journal of computational biology: a journal of computational molecular cell biology, vol. 31, iss. 6, no. 6, pp. 549-563, 2024.
@article{vonLoehneysen:24:phylogenetic,
title = {Phylogenetic and chemical probing information as soft constraints in RNA secondary structure prediction.},
author = {Sarah Löhneysen and Thomas Spicher and Yuliia Varenyk and Hua-Ting Yao and Ronny Lorenz and Ivo Hofacker and Peter F Stadler},
url = {https://pubmed.ncbi.nlm.nih.gov/38935442/},
doi = {10.1089/cmb.2024.0519},
year = {2024},
date = {2024-06-01},
journal = {Journal of computational biology: a journal of computational molecular cell biology},
volume = {31},
number = {6},
issue = {6},
pages = {549-563},
keywords = {},
pubstate = {ppublish},
tppubtype = {article}
}
Günther, Kerstin; Ehrhardt, Christina; Werz, Oliver; Jordan, Paul M
Protocol for lipid mediator profiling and phenotyping of human M1-and M2-monocyte-derived macrophages during host-pathogen interactions Journal Article
In: STAR protocols, vol. 5, no. 3, pp. 103142, 2024.
@article{Guenther:24,
title = {Protocol for lipid mediator profiling and phenotyping of human M1-and M2-monocyte-derived macrophages during host-pathogen interactions},
author = {Kerstin Günther and Christina Ehrhardt and Oliver Werz and Paul M Jordan},
year = {2024},
date = {2024-01-01},
journal = {STAR protocols},
volume = {5},
number = {3},
pages = {103142},
publisher = {Elsevier},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Jordan, Paul M; Günther, Kerstin; Nischang, Vivien; Ning, Yuping; Deinhardt-Emmer, Stefanie; Ehrhardt, Christina; Werz, Oliver
Influenza A virus selectively elevates prostaglandin E2 formation in pro-resolving macrophages Journal Article
In: iScience, vol. 27, iss. 1, no. 1, pp. 108775, 2024.
@article{Jordan:24,
title = {Influenza A virus selectively elevates prostaglandin E2 formation in pro-resolving macrophages},
author = {Paul M Jordan and Kerstin Günther and Vivien Nischang and Yuping Ning and Stefanie Deinhardt-Emmer and Christina Ehrhardt and Oliver Werz},
url = {https://pubmed.ncbi.nlm.nih.gov/38261967/},
doi = {10.1016/j.isci.2023.108775},
year = {2024},
date = {2024-01-01},
journal = {iScience},
volume = {27},
number = {1},
issue = {1},
pages = {108775},
publisher = {Elsevier},
keywords = {},
pubstate = {epublish},
tppubtype = {article}
}
2023
Klapproth, Christopher; Zötzsche, Siegfried; Kühnl, Felix; Fallmann, Jörg; Stadler, Peter F; Findeiß, Sven
Tailored machine learning models for functional RNA detection in genome-wide screens. Journal Article
In: NAR Genom Bioinform, vol. 5, iss. 3, no. 3, pp. lqad072, 2023.
@article{Klapproth:23,
title = {Tailored machine learning models for functional RNA detection in genome-wide screens.},
author = {Christopher Klapproth and Siegfried Zötzsche and Felix Kühnl and Jörg Fallmann and Peter F Stadler and Sven Findeiß},
url = {https://pubmed.ncbi.nlm.nih.gov/37608800/},
doi = {10.1093/nargab/lqad072},
year = {2023},
date = {2023-09-01},
journal = {NAR Genom Bioinform},
volume = {5},
number = {3},
issue = {3},
pages = {lqad072},
keywords = {},
pubstate = {epublish},
tppubtype = {article}
}
2022
Jordan, Paul M; Werz, Oliver
Specialized pro-resolving mediators: Biosynthesis and biological role in bacterial infections Journal Article
In: The FEBS journal, vol. 289, no. 14, pp. 4212–4227, 2022.
@article{Jordan:22,
title = {Specialized pro-resolving mediators: Biosynthesis and biological role in bacterial infections},
author = {Paul M Jordan and Oliver Werz},
year = {2022},
date = {2022-01-01},
journal = {The FEBS journal},
volume = {289},
number = {14},
pages = {4212–4227},
publisher = {Wiley Online Library},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2020
Jordan, Paul M; Gerstmeier, Jana; Pace, Simona; Bilancia, Rossella; Rao, Zhigang; Börner, Friedemann; Miek, Laura; Gutiérrez-Gutiérrez, Óscar; Arakandy, Vandana; Rossi, Antonietta; others,
Staphylococcus aureus-derived α-hemolysin evokes generation of specialized pro-resolving mediators promoting inflammation resolution Journal Article
In: Cell Rep, vol. 33, no. 2, 2020.
@article{Jordan:20,
title = {Staphylococcus aureus-derived α-hemolysin evokes generation of specialized pro-resolving mediators promoting inflammation resolution},
author = {Paul M Jordan and Jana Gerstmeier and Simona Pace and Rossella Bilancia and Zhigang Rao and Friedemann Börner and Laura Miek and Óscar Gutiérrez-Gutiérrez and Vandana Arakandy and Antonietta Rossi and others},
year = {2020},
date = {2020-01-01},
journal = {Cell Rep},
volume = {33},
number = {2},
publisher = {Elsevier},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2011
Lorenz, Ronny; Bernhart, Stephan H; Siederdissen, Christian Höner Zu; Tafer, Hakim; Flamm, Christoph; Stadler, Peter F; Hofacker, Ivo L
ViennaRNA Package 2.0 Journal Article
In: Alg Mol Biol, vol. 6, pp. 26, 2011.
@article{Lorenz:11,
title = {ViennaRNA Package 2.0},
author = {Ronny Lorenz and Stephan H Bernhart and Christian Höner Zu Siederdissen and Hakim Tafer and Christoph Flamm and Peter F Stadler and Ivo L Hofacker},
url = {https://pubmed.ncbi.nlm.nih.gov/22115189/},
doi = {10.1186/1748-7188-6-26},
year = {2011},
date = {2011-11-01},
journal = {Alg Mol Biol},
volume = {6},
pages = {26},
keywords = {},
pubstate = {epublish},
tppubtype = {article}
}
2004
Thurner, Caroline; Witwer, Christine; Hofacker, Ivo; Stadler, Peter F.
Conserved RNA secondary structures in emphFlaviviridae genomes Journal Article
In: J Gen Virol, vol. 85, pp. 1113-1124, 2004.
@article{Thurner:04a,
title = {Conserved RNA secondary structures in emphFlaviviridae genomes},
author = {Caroline Thurner and Christine Witwer and Ivo Hofacker and Peter F. Stadler},
doi = {10.1099/vir.0.19462-0},
year = {2004},
date = {2004-01-01},
journal = {J Gen Virol},
volume = {85},
pages = {1113-1124},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
1998
Hofacker, I L; Fekete, M; Flamm, C; Huynen, M A; Rauscher, S; Stolorz, P E; Stadler, Peter F.
Automatic detection of conserved RNA structure elements in complete RNA virus genomes Journal Article
In: Nucleic Acids Res, vol. 26, iss. 16, pp. 3825–3836, 1998, (Santa Fe Institute Preprint 98-03-020).
@article{Hofacker:98,
title = {Automatic detection of conserved RNA structure elements in complete RNA virus genomes},
author = {I L Hofacker and M Fekete and C Flamm and M A Huynen and S Rauscher and P E Stolorz and Peter F. Stadler},
url = {https://pubmed.ncbi.nlm.nih.gov/9685502/},
doi = {10.1093/nar/26.16.3825},
year = {1998},
date = {1998-08-01},
journal = {Nucleic Acids Res},
volume = {26},
issue = {16},
pages = {3825–3836},
note = {Santa Fe Institute Preprint 98-03-020},
keywords = {},
pubstate = {ppublish},
tppubtype = {article}
}



