
C04: Raman spectroscopy: A tool for host response and virus characterisation

Projects of the CRC 1768

C04: Raman spectroscopy: A tool for host response and virus characterisation
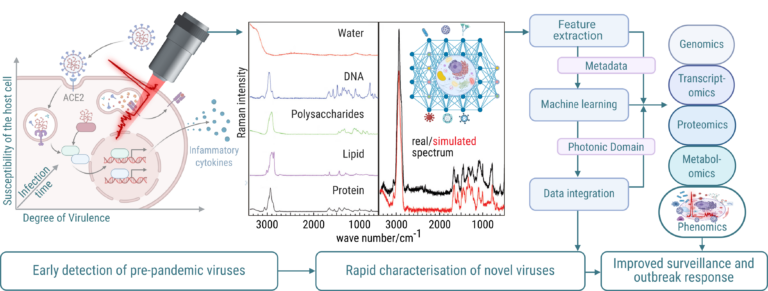
Its rapid, label-free measurements provide a versatile tool for investigating the changes in a cell’s morphochemical pattern, which controls many key processes during infection. Modern Raman-based analyses leverage machine learning to integrate spectral information within the biological context, revealing biochemical changes that drive phenotypic adaptations during infection – opening new dimensions for rapidly understanding underlying virus tropism. Building on established Raman spectroscopic fingerprinting of bacterial infections, we aim to extend this powerful technique to virus systems, offering rapid, scalable, and reproducible insights essential for pandemic preparedness, early detection of emerging pathogens, and zoonotic disease surveillance. By leveraging advanced Raman spectroscopic workflows combined with machine learning algorithms, the project aims to translate complex spectral data into actionable phenotypic insights. To ensure reproducibility and comparability, we will establish standardised and rigorously validated sample preparation protocols optimised specifically for Raman analysis, providing a robust foundation for elucidating virus infection cycles. Crucially, the tool we develop will systematically integrate Raman spectroscopy data with multiomics datasets (genomic, transcriptomic, proteomic, and metabolomic) for the analysis of virus interactions across different host systems by investigating infections with the eukaryotic virus SARS-CoV-2 and the vibriophage N4.
Project Overview
- We will utilise SARS-CoV-2 variants with varying pathogenicity and transmissibility (from virulent to attenuated to single cycle) in conjunction with host cells exhibiting different susceptibility levels, in order to comprehensively develop and characterise the tool’s output. This will establish a robust baseline for characterising virus-host interactions applicable to the characterisation of new viruses.
- We will then transfer these technological findings and optimised protocols to the investigation of Vibrio cholerae under vibriophage N4 infection and Klebsiella spp. under specific phage infection, expanding the platform’s applicability to prokaryotic systems. We will perform a comparative analysis of the initial results obtained from eukaryotic virus infection (SARS-CoV-2) and prokaryotic vibriophage N4 infection to identify conserved and divergent mechanisms of host-pathogen interaction, providing a broader understanding of fundamental principles governing these processes. This comparative approach will allow us to identify both universal and specific responses to infection across different domains of life.
- Building on our established expertise in sample preparation for Raman spectroscopic investigation of biological samples, we will investigate label-free phenotypic characterisation of intact virus particles.
- Tool to be developed: We will develop a fully integrated Raman spectroscopy platform optimised for virus research – from sample preparation to machine learning-guided data interpretation – enabling label-free, high-resolution, and standardised characterisation of virus-host systems. The tool translates virus insights captured in the photonic domain to provide relevant alerts and insights for decision systems.
Hypothesis enabled by the proposed tool: The proposed tool enables the hypothesis that Raman spectral fingerprints of virus particles and infected host cells capture distinct molecular signatures reflecting infection status, strainspecific virus effects, and host cell reprogramming, bridging genotypic and phenotypic insights across eukaryotic and prokaryotic systems.
Overarching CRC goals: Our project C04 establishes a Raman spectroscopy-based, label-free phenotyping platform with standardised workflows and ML to capture morphochemical host-response fingerprints and virion features, enabling rapid, reproducible description of virus-host interactions (G1, G3). Integrating spectra with multi-omics, the tool yields calibrated predictors of tropism, pathogenicity, and virulence for early triage and pandemic preparedness (G4).
Work Packages (WP):
- WP 1: Raman spectroscopy platform optimised for virus research in the eukaryotic domain (Beer/Popp)
- WP 2: Raman spectroscopy platform optimised for virus research in the prokaryotic domain (Popp)
- WP 3: Technological benchmark for the label-free characterisation of virus particles (Popp/Beer)
Team Members
2025
Ramoji, Anuradha; Baumbach, Philipp; Ryabchykov, Oleg; Pistiki, Aikaterini; Rueger, Jan; Pinzon, David Vasquez; Silge, Anja; Deinhardt-Emmer, Stefanie; Schie, Iwan W; Weber, Karina; others,
Raman Spectroscopy Can Identify Acute and Persistent Biochemical Changes in Leukocytes From Patients With COVID-19 and Non-COVID-19-Associated Sepsis Journal Article
In: Biotechnology Journal, vol. 20, no. 9, pp. e70105, 2025.
@article{ramoji2025raman,
title = {Raman Spectroscopy Can Identify Acute and Persistent Biochemical Changes in Leukocytes From Patients With COVID-19 and Non-COVID-19-Associated Sepsis},
author = {Anuradha Ramoji and Philipp Baumbach and Oleg Ryabchykov and Aikaterini Pistiki and Jan Rueger and David Vasquez Pinzon and Anja Silge and Stefanie Deinhardt-Emmer and Iwan W Schie and Karina Weber and others},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Biotechnology Journal},
volume = {20},
number = {9},
pages = {e70105},
publisher = {Wiley Online Library},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2021
Wernike, Kerstin; Reimann, Ilona; Banyard, Ashley C; Kraatz, Franziska; Rocca, S Anna La; Hoffmann, Bernd; McGowan, Sarah; Hechinger, Silke; Choudhury, Bhudipa; Aebischer, Andrea; others,
High genetic variability of Schmallenberg virus M-segment leads to efficient immune escape from neutralizing antibodies Journal Article
In: PLoS pathogens, vol. 17, no. 1, pp. e1009247, 2021.
@article{wernike2021high,
title = {High genetic variability of Schmallenberg virus M-segment leads to efficient immune escape from neutralizing antibodies},
author = {Kerstin Wernike and Ilona Reimann and Ashley C Banyard and Franziska Kraatz and S Anna La Rocca and Bernd Hoffmann and Sarah McGowan and Silke Hechinger and Bhudipa Choudhury and Andrea Aebischer and others},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {PLoS pathogens},
volume = {17},
number = {1},
pages = {e1009247},
publisher = {Public Library of Science San Francisco, CA USA},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Guo, Shuxia; Popp, Jürgen; Bocklitz, Thomas
Chemometric analysis in Raman spectroscopy from experimental design to machine learning–based modeling Journal Article
In: Nature protocols, vol. 16, no. 12, pp. 5426–5459, 2021.
@article{guo2021chemometric,
title = {Chemometric analysis in Raman spectroscopy from experimental design to machine learning–based modeling},
author = {Shuxia Guo and Jürgen Popp and Thomas Bocklitz},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Nature protocols},
volume = {16},
number = {12},
pages = {5426–5459},
publisher = {Nature Publishing Group UK London},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2020
Dalmann, Anja; Reimann, Ilona; Wernike, Kerstin; Beer, Martin
Autonomously replicating RNAs of Bungowannah pestivirus: ERNS is not essential for the generation of infectious particles Journal Article
In: Journal of Virology, vol. 94, no. 14, pp. 10–1128, 2020.
@article{dalmann2020autonomously,
title = {Autonomously replicating RNAs of Bungowannah pestivirus: ERNS is not essential for the generation of infectious particles},
author = {Anja Dalmann and Ilona Reimann and Kerstin Wernike and Martin Beer},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {Journal of Virology},
volume = {94},
number = {14},
pages = {10–1128},
publisher = {American Society for Microbiology 1752 N St., NW, Washington, DC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Thamamongood, Thiprampai; Aebischer, Andrea; Wagner, Valentina; Chang, Max W; Elling, Roland; Benner, Christopher; García-Sastre, Adolfo; Kochs, Georg; Beer, Martin; Schwemmle, Martin
A genome-wide CRISPR-Cas9 screen reveals the requirement of host cell sulfation for Schmallenberg virus infection Journal Article
In: Journal of Virology, vol. 94, no. 17, pp. 10–1128, 2020.
@article{thamamongood2020genome,
title = {A genome-wide CRISPR-Cas9 screen reveals the requirement of host cell sulfation for Schmallenberg virus infection},
author = {Thiprampai Thamamongood and Andrea Aebischer and Valentina Wagner and Max W Chang and Roland Elling and Christopher Benner and Adolfo García-Sastre and Georg Kochs and Martin Beer and Martin Schwemmle},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {Journal of Virology},
volume = {94},
number = {17},
pages = {10–1128},
publisher = {American Society for Microbiology 1752 N St., NW, Washington, DC},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Arend, Natalie; Pittner, Angelina; Ramoji, Anuradha; Mondol, Abdullah S; Dahms, Marcel; Rüger, Jan; Kurzai, Oliver; Schie, Iwan W; Bauer, Michael; Popp, Jürgen; others,
Detection and differentiation of bacterial and fungal infection of neutrophils from peripheral blood using Raman spectroscopy Journal Article
In: Analytical chemistry, vol. 92, no. 15, pp. 10560–10568, 2020.
@article{arend2020detection,
title = {Detection and differentiation of bacterial and fungal infection of neutrophils from peripheral blood using Raman spectroscopy},
author = {Natalie Arend and Angelina Pittner and Anuradha Ramoji and Abdullah S Mondol and Marcel Dahms and Jan Rüger and Oliver Kurzai and Iwan W Schie and Michael Bauer and Jürgen Popp and others},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {Analytical chemistry},
volume = {92},
number = {15},
pages = {10560–10568},
publisher = {ACS Publications},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Deckert, Volker; Deckert-Gaudig, Tanja; Cialla-May, Dana; Popp, Jürgen; Zell, Roland; Deinhard-Emmer, Stefanie; Sokolov, Alexei V; Yi, Zhenhuan; Scully, Marlan O
Laser spectroscopic technique for direct identification of a single virus I: FASTER CARS Journal Article
In: Proceedings of the National Academy of Sciences, vol. 117, no. 45, pp. 27820–27824, 2020.
@article{deckert2020laserb,
title = {Laser spectroscopic technique for direct identification of a single virus I: FASTER CARS},
author = {Volker Deckert and Tanja Deckert-Gaudig and Dana Cialla-May and Jürgen Popp and Roland Zell and Stefanie Deinhard-Emmer and Alexei V Sokolov and Zhenhuan Yi and Marlan O Scully},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {Proceedings of the National Academy of Sciences},
volume = {117},
number = {45},
pages = {27820–27824},
publisher = {National Academy of Sciences},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2018
Kirchhoff, Johanna; Glaser, Uwe; Bohnert, Jürgen A; Pletz, Mathias W; Popp, Jürgen; Neugebauer, Ute
Simple ciprofloxacin resistance test and determination of minimal inhibitory concentration within 2 h using Raman spectroscopy Journal Article
In: Analytical chemistry, vol. 90, no. 3, pp. 1811–1818, 2018.
@article{kirchhoff2018simple,
title = {Simple ciprofloxacin resistance test and determination of minimal inhibitory concentration within 2 h using Raman spectroscopy},
author = {Johanna Kirchhoff and Uwe Glaser and Jürgen A Bohnert and Mathias W Pletz and Jürgen Popp and Ute Neugebauer},
year = {2018},
date = {2018-01-01},
urldate = {2018-01-01},
journal = {Analytical chemistry},
volume = {90},
number = {3},
pages = {1811–1818},
publisher = {ACS Publications},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2015
Hoffmann, Bernd; Tappe, Dennis; Höper, Dirk; Herden, Christiane; Boldt, Annemarie; Mawrin, Christian; Niederstraßer, Olaf; Müller, Tobias; Jenckel, Maria; Grinten, Elisabeth; others,
A variegated squirrel bornavirus associated with fatal human encephalitis Journal Article
In: New England Journal of Medicine, vol. 373, no. 2, pp. 154–162, 2015.
@article{hoffmann2015variegated,
title = {A variegated squirrel bornavirus associated with fatal human encephalitis},
author = {Bernd Hoffmann and Dennis Tappe and Dirk Höper and Christiane Herden and Annemarie Boldt and Christian Mawrin and Olaf Niederstraßer and Tobias Müller and Maria Jenckel and Elisabeth Grinten and others},
year = {2015},
date = {2015-01-01},
urldate = {2015-01-01},
journal = {New England Journal of Medicine},
volume = {373},
number = {2},
pages = {154–162},
publisher = {Mass Medical Soc},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2012
Hoffmann, Bernd; Scheuch, Matthias; Höper, Dirk; Jungblut, Ralf; Holsteg, Mark; Schirrmeier, Horst; Eschbaumer, Michael; Goller, Katja V; Wernike, Kerstin; Fischer, Melina; others,
Novel orthobunyavirus in cattle, Europe, 2011 Journal Article
In: Emerging infectious diseases, vol. 18, no. 3, pp. 469, 2012.
@article{hoffmann2012novel,
title = {Novel orthobunyavirus in cattle, Europe, 2011},
author = {Bernd Hoffmann and Matthias Scheuch and Dirk Höper and Ralf Jungblut and Mark Holsteg and Horst Schirrmeier and Michael Eschbaumer and Katja V Goller and Kerstin Wernike and Melina Fischer and others},
year = {2012},
date = {2012-01-01},
urldate = {2012-01-01},
journal = {Emerging infectious diseases},
volume = {18},
number = {3},
pages = {469},
keywords = {},
pubstate = {published},
tppubtype = {article}
}



