
C02: Probe microscopy-based functional tracking of respiratory viruses to identify virus tropism

Projects of the CRC 1768

C02: Probe microscopy-based functional tracking of respiratory viruses to identify virus tropism
We aim to characterise virus tropism by identifying permissivecelltypesthatsupportcompletevirusrepli cation by using Tip-enhanced Raman Spectroscopy (TERS). TERS integrates atomic force microscopy (AFM) and near-field optical recordings with Raman spectroscopy, enabling direct, label-free characterisa tion of molecular organisation and thus visualisation of virus entry and exit at nanometer resolution.
As a result, tropism ultimately defines whether productive replication occurs and thereby shapes virus pathogenesis, disease progression, and transmission. However, inferring tropism from virus properties alone remains difficult and typically requires elaborate infection experiments.
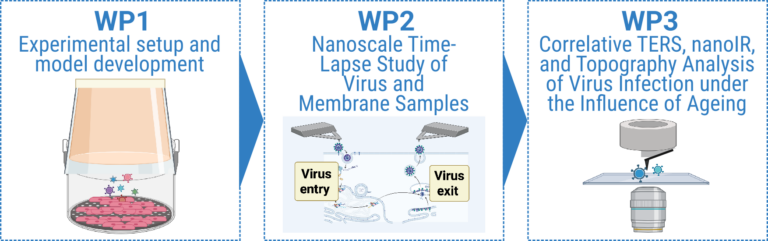
In this project, TERS will be employed to resolve virus entry and exit strategies with unprecedented spatial detail, Fig. C02.1. Beyondimaging, TERSprovidesmolecularfingerprintsandresolvesnanoscalesurfacestructures, thereby revealing chemical and conformational changes that accompany virus-cell interactions. Time-resolved nanoscale studies will further allow us to monitor replication at the level of single infection events and to detect heterogeneous infection patterns that are masked in bulk analyses. By combining structural and temporal information, we aim to precisely definethehostcellfactorsandphysiologicalstatesthatpermitorrestrictvirus replication, thus contributing directly to research goals G3 and G4. Our approach represents a novel, rapid, label-free, and phenotypic method to define virus tropism. In addition to TERS, we will employ nanoscale infrared spectroscopy (nanoIR), a technique that combines AFM with IR absorption to provide chemical composition and conformational information at nanometer resolution. The combination of nanoIR and TERS will yield complementary spectroscopic data to unravel structural changes during virus-cell interactions, potentially down to amino acid resolution. To further dissect the influence of host determinants on virus tropism, we will additionally apply a cellular senescence model.
Project Overview
Objectives
a. Objective1: Developmentofavirus-compatiblehigh-resolutioninfectionmodel.
b. Objective 2: Nanoscale visualisation of virus entry and budding dynamics.
c. Objective 3: Investigating the impact of ageing on virus tropism with anomechanic and nano-spectroscopic tools.
- Tool to be developed: A method to investigate single virus-membrane interactions, combining atomic force microscopy and(Raman)spectroscopy. Thesestructurally sensitive nanoscale spectroscopy techniques, particularly Raman and infrared, will lead to a fast and label-free photonic tool to identify tropism.
Hypothesis enabled by the proposed tool: We hypothesise that age-related changes in the host tissue microenvi ronment act as selective pressures that drive adaptive shifts in virus tropism, ultimately altering virus replication dynamics, tissue specificity, and pathogenic potential.
Overarching CRC goals: Our project C02 leverages AFM-TERS and complementary nanoIR to track single-virus entry, replication, and budding with nanometer, time-resolved molecular fingerprints, defining host-cell determinants and physiological states that permit productive replication and deriving generalisable rules of tropism across contexts (G1, G3). Integrating senescence models and phenotypic spectra enables predictive assessment of tissue/cell-type permissivity, and infection potential, supporting tropism forecasting in aged versus non-aged environments (G4).
Work Packages (WP):
- WP 1: Experimental setup and model development (Deckert/Deinhardt-Emmer)
- WP 2: Nanoscale time-lapse study of virus and membrane samples (Deckert/Deinhardt-Emmer)
- WP 3: Correlative TERS, nanoIR, and topography analysis of virus infection (Deckert/Deinhardt-Emmer)
Team Members
2025
Hornung, Franziska; SureshKumar, Harini K; Klement, Laura; Reisser, Yasmina; Wernike, Christoph; Nischang, Vivien; Jordan, Paul M; Werz, Oliver; Hoffmann, Carsten; Löffler, Bettina; others,
High-fat diet impairs microbial metabolite production and aggravates influenza A infection Journal Article
In: Cell Communication and Signaling, vol. 23, no. 1, pp. 359, 2025.
@article{hornung2025highb,
title = {High-fat diet impairs microbial metabolite production and aggravates influenza A infection},
author = {Franziska Hornung and Harini K SureshKumar and Laura Klement and Yasmina Reisser and Christoph Wernike and Vivien Nischang and Paul M Jordan and Oliver Werz and Carsten Hoffmann and Bettina Löffler and others},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Cell Communication and Signaling},
volume = {23},
number = {1},
pages = {359},
publisher = {Springer},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Deinhardt-Emmer, Stefanie; Chousterman, Benjamin G; Schefold, Joerg C; Flohé, Stefanie B; Skirecki, Tomasz; Kox, Matthijs; Winkler, Martin S; Cossarizza, Andrea; Wiersinga, W Joost; Poll, Tom; others,
Sepsis in patients who are immunocompromised: diagnostic challenges and future therapies Journal Article
In: The Lancet Respiratory Medicine, vol. 13, no. 7, pp. 623–637, 2025.
@article{deinhardt2025sepsis,
title = {Sepsis in patients who are immunocompromised: diagnostic challenges and future therapies},
author = {Stefanie Deinhardt-Emmer and Benjamin G Chousterman and Joerg C Schefold and Stefanie B Flohé and Tomasz Skirecki and Matthijs Kox and Martin S Winkler and Andrea Cossarizza and W Joost Wiersinga and Tom Poll and others},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {The Lancet Respiratory Medicine},
volume = {13},
number = {7},
pages = {623–637},
publisher = {Elsevier},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Höppener, Christiane; Sobotta, Fabian H; Hoeppener, Stephanie; Deckert, Volker; Brendel, Johannes C
Unraveling the Hierarchical Self-Assembly of Amphiphilic Block Copolymer-Peptide Conjugates by Tip-Enhanced Raman Spectroscopy Journal Article
In: Small, pp. 2502157, 2025.
@article{hoppener2025unraveling,
title = {Unraveling the Hierarchical Self-Assembly of Amphiphilic Block Copolymer-Peptide Conjugates by Tip-Enhanced Raman Spectroscopy},
author = {Christiane Höppener and Fabian H Sobotta and Stephanie Hoeppener and Volker Deckert and Johannes C Brendel},
year = {2025},
date = {2025-01-01},
urldate = {2025-01-01},
journal = {Small},
pages = {2502157},
publisher = {Wiley Online Library},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2024
Höppener, Christiane; Aizpurua, Javier; Chen, Huan; Gräfe, Stefanie; Jorio, Ado; Kupfer, Stephan; Zhang, Zhenglong; Deckert, Volker
Tip-enhanced Raman scattering Journal Article
In: Nature Reviews Methods Primers, vol. 4, no. 1, pp. 47, 2024.
@article{hoppener2024tip,
title = {Tip-enhanced Raman scattering},
author = {Christiane Höppener and Javier Aizpurua and Huan Chen and Stefanie Gräfe and Ado Jorio and Stephan Kupfer and Zhenglong Zhang and Volker Deckert},
year = {2024},
date = {2024-01-01},
urldate = {2024-01-01},
journal = {Nature Reviews Methods Primers},
volume = {4},
number = {1},
pages = {47},
publisher = {Nature Publishing Group UK London},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2023
Schulz, Luise; Hornung, Franziska; Häder, Antje; Radosa, Lukáš; Brakhage, Axel A; Löffler, Bettina; Deinhardt-Emmer, Stefanie
Influenza virus-induced paracrine cellular senescence of the lung contributes to enhanced viral load Journal Article
In: Aging and disease, vol. 14, no. 4, pp. 1331, 2023.
@article{schulz2023influenza,
title = {Influenza virus-induced paracrine cellular senescence of the lung contributes to enhanced viral load},
author = {Luise Schulz and Franziska Hornung and Antje Häder and Lukáš Radosa and Axel A Brakhage and Bettina Löffler and Stefanie Deinhardt-Emmer},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {Aging and disease},
volume = {14},
number = {4},
pages = {1331},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Deckert-Gaudig, Tanja; Yao, Xiaobin; Darussalam, Erwan; Hornung, Franziska; Carravilla, Pablo; Zhao, Ziliang; Rezaei, Kourosh; Eggeling, Christian; Deinhardt-Emmer, Stefanie; Deckert, Volker
Identification of RNA-containing virus particles using a triple correlative morphological and microscopic approach Journal Article
In: 2023.
@article{deckert2023identification,
title = {Identification of RNA-containing virus particles using a triple correlative morphological and microscopic approach},
author = {Tanja Deckert-Gaudig and Xiaobin Yao and Erwan Darussalam and Franziska Hornung and Pablo Carravilla and Ziliang Zhao and Kourosh Rezaei and Christian Eggeling and Stefanie Deinhardt-Emmer and Volker Deckert},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Höppener, Christiane; Elter, Johanna K; Schacher, Felix H; Deckert, Volker
Inside block copolymer micelles—tracing interfacial influences on crosslinking efficiency in nanoscale confined spaces Journal Article
In: Small, vol. 19, no. 20, pp. 2206451, 2023.
@article{hoppener2023inside,
title = {Inside block copolymer micelles—tracing interfacial influences on crosslinking efficiency in nanoscale confined spaces},
author = {Christiane Höppener and Johanna K Elter and Felix H Schacher and Volker Deckert},
year = {2023},
date = {2023-01-01},
urldate = {2023-01-01},
journal = {Small},
volume = {19},
number = {20},
pages = {2206451},
publisher = {Wiley Online Library},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2021
Deinhardt-Emmer, Stefanie; Wittschieber, Daniel; Sanft, Juliane; Kleemann, Sandra; Elschner, Stefan; Haupt, Karoline Frieda; Vau, Vanessa; Häring, Clio; Rödel, Jürgen; Henke, Andreas; others,
Early postmortem mapping of SARS-CoV-2 RNA in patients with COVID-19 and the correlation with tissue damage Journal Article
In: Elife, vol. 10, pp. e60361, 2021.
@article{deinhardt2021early,
title = {Early postmortem mapping of SARS-CoV-2 RNA in patients with COVID-19 and the correlation with tissue damage},
author = {Stefanie Deinhardt-Emmer and Daniel Wittschieber and Juliane Sanft and Sandra Kleemann and Stefan Elschner and Karoline Frieda Haupt and Vanessa Vau and Clio Häring and Jürgen Rödel and Andreas Henke and others},
year = {2021},
date = {2021-01-01},
urldate = {2021-01-01},
journal = {Elife},
volume = {10},
pages = {e60361},
publisher = {eLife Sciences Publications, Ltd},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2020
Deckert, Volker; Deckert-Gaudig, Tanja; Cialla-May, Dana; Popp, Jürgen; Zell, Roland; Deinhard-Emmer, Stefanie; Sokolov, Alexei V; Yi, Zhenhuan; Scully, Marlan O
Laser spectroscopic technique for direct identification of a single virus I: FASTER CARS Journal Article
In: Proceedings of the National Academy of Sciences, vol. 117, no. 45, pp. 27820–27824, 2020.
@article{deckert2020laser,
title = {Laser spectroscopic technique for direct identification of a single virus I: FASTER CARS},
author = {Volker Deckert and Tanja Deckert-Gaudig and Dana Cialla-May and Jürgen Popp and Roland Zell and Stefanie Deinhard-Emmer and Alexei V Sokolov and Zhenhuan Yi and Marlan O Scully},
year = {2020},
date = {2020-01-01},
urldate = {2020-01-01},
journal = {Proceedings of the National Academy of Sciences},
volume = {117},
number = {45},
pages = {27820–27824},
publisher = {National Academy of Sciences},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
2015
Olschewski, Konstanze; Kämmer, Evelyn; Stöckel, Stephan; Bocklitz, Thomas; Deckert-Gaudig, Tanja; Zell, Roland; Cialla-May, Dana; Weber, Karina; Deckert, Volker; Popp, Jürgen
A manual and an automatic TERS based virus discrimination Journal Article
In: Nanoscale, vol. 7, no. 10, pp. 4545–4552, 2015.
@article{olschewski2015manual,
title = {A manual and an automatic TERS based virus discrimination},
author = {Konstanze Olschewski and Evelyn Kämmer and Stephan Stöckel and Thomas Bocklitz and Tanja Deckert-Gaudig and Roland Zell and Dana Cialla-May and Karina Weber and Volker Deckert and Jürgen Popp},
year = {2015},
date = {2015-01-01},
urldate = {2015-01-01},
journal = {Nanoscale},
volume = {7},
number = {10},
pages = {4545–4552},
publisher = {Royal Society of Chemistry},
keywords = {},
pubstate = {published},
tppubtype = {article}
}


